Tutorial 1. Simulation of 1D NMR line shape with IDAP
by Evgenii Kovrigin, 05/17/2011
Here I am showing coding steps to necessary to simulate 1D NMR line shapes for a simple R+L=RL process (U model). In this model we observe signals from R and RL while L is NMR-invisible (unlabeled with NMR-active isotopes). I will set some parameters for the U model and prepare a single 1D spectrum. I show how to export data and how to use both analytical and numeric solutions of the model.
I only intend to do simulation of a signal here. For fitting (later) we should use functionality of the TotalFit class
Contents
- Clean up workspace
- Set up model parameters for the 1D NMR line shape
- Create NMR line shape dataset
- Model, solver and solver options
- Simulate ideal data
- Simulate data with synthetic random noise
- Save a dataset object
- Example of using numeric solution of the models
- Estimation of the NMR line parameters
- Conclusions
Clean up workspace
(to avoid unwanted effects of old variable assignments)
close all clear all
Set up model parameters for the 1D NMR line shape
I will be building a setup structure to be able to handle all these parameters as a single piece (therefore--exclusively for convenience)
setup.Rtotal = 1e-3; % concentration of receptor, mol setup.LRratio = 0.75; % ratio of the total concentration of ligand to receptor setup.Log10_K_A = log10(1e5); % equilibrium binding constant, 1/mol, in logarithmic form setup.k_2_A = 10; % dissociation rate constant of the complex, 1/s setup.w0_1 = 0; % NMR frequency of the free receptor R, 1/s setup.w0_2 = 200; % NMR frequency of the bound complex RL, 1/s setup.FWHH_1 = 20; % line width at half height of the peak of R, 1/s setup.FWHH_2 = 20; % line width at half height of the peak of RL, 1/s setup.ScaleFactor = 1; % a multiplier for spectral amplitude (used only when fitting data)
Create NMR line shape dataset
Create data object for 1D NMR line shapes and list all models defined for this data type.
test1=NMRLineShapes1D('Simulation','Test_1'); % create a dataset object test1.list_models() % show existing models
ans = Models defined in the object "Test_1" of type "1D NMR Line shapes" ----------------------------------------------------------- Model 1: "Lorentzian" Model description "Lorentzian function, analytical" Model handle: line_shape_equations_1D.lorentzian Current solver: analytical Model parameters: 1: Intensity 2: dw 3: w0 Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor Model 3: "U-model-LRcorrection" Model description "1D NMR line shape for the U model, no temperature/field dependence, with L/R correction, analytical" Model handle: line_shape_equations_1D.U_model_1D_LRcorrection Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor 10: LRcorrection Model 4: "U-model-LRcorr-Jdoublet" Model description "1D NMR line shape for the U model split into a doublet with a constant J (1/s), no temperature/field dependence, with L/R correction, analytical" Model handle: line_shape_equations_1D.U_model_1D_LRcorr_doublet Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor 10: LRcorrection 11: Jconst Model 5: "U_model_1D_LR_LB_SF" Model description "1D NMR line shape, U model, no T/B0 dep, with scaling factor, L/R correction and linear baseline, analytical" Model handle: line_shape_equations_1D.U_model_1D_LR_LB_SF Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor 10: LRcorrection 11: LB_a 12: LB_b Model 6: "U_R-model" Model description "1D NMR line shape for the U-R model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_R_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: log10(K_B) 5: k_2_A 6: k_2_B 7: w0_R 8: w0_Rstar 9: w0_RL 10: FWHH_R 11: FWHH_Rstar 12: FWHH_RL 13: ScaleFactor Model 7: "U_R2-model" Model description "1D NMR line shape for the U-R2 model, no temperature/field dependence, fminbnd solver" Model handle: line_shape_equations_1D.U_R2_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: log10(K_B) 5: k_2_A 6: k_2_B 7: w0_R 8: w0_R2 9: w0_RL 10: FWHH_R 11: FWHH_R2 12: FWHH_RL 13: ScaleFactor Model 8: "U_RL-model" Model description "1D NMR line shape for the U-RL model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_RL_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: log10(K_B) 5: k_2_A 6: k_2_B 7: w0_R 8: w0_RL 9: w0_RLstar 10: FWHH_R 11: FWHH_RL 12: FWHH_RLstar 13: ScaleFactor Model 9: "U_R2L2-model" Model description "1D NMR line shape for the U-R2L2 model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_R2L2_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: log10(K_B) 5: k_2_A 6: k_2_B 7: w0_R 8: w0_RL 9: w0_R2L2 10: FWHH_R 11: FWHH_RL 12: FWHH_R2L2 13: ScaleFactor Model 10: "U_R_L_RL-a-model" Model description "1D NMR line shape for the U-R-L-RL_a model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_R_L_RL_a_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A1) 4: log10(K_B1) 5: log10(K_B2) 6: log10(K_B3) 7: k_2_A1 8: k_2_B1 9: k_2_B2 10: k_2_B3 11: w0_R 12: w0_Rstar 13: w0_RL 14: w0_RLstar 15: FWHH_R 16: FWHH_Rstar 17: FWHH_RL 18: FWHH_RLstar 19: ScaleFactor
Model, solver and solver options
First, we specify what model we want to use. For models that do not have analytical solutions we should specify a solver and its options. U model is soluble analytically so none of the options is required.
test1.set_active_model('U-model', 'analytical', 'no options') % select a model, solver and its options test1.show_active_model()
ans = Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor
Simulate ideal data
Set range for X to extend beyond resonances
w_padding=100; % extend X axis by this amount beyond resonance positions w_min= min([setup.w0_2,setup.w0_1])-w_padding; w_max= max([setup.w0_2,setup.w0_1])+w_padding; datapoints=50; % Number of simulated 'experimental' data points test1.set_X(linspace(w_min, w_max, datapoints));
Calculate ideal data: set noise RMSD to 0 for both X and Y in this example:
X_RMSD=0; Y_RMSD=0;
Collect all parameters into a vector in the same order as model requires
parameters=[ setup.Rtotal setup.LRratio setup.Log10_K_A setup.k_2_A ... setup.w0_1 setup.w0_2 setup.FWHH_1 setup.FWHH_2 ... setup.ScaleFactor ];
Compute simulated line shape
tic;
test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD);
toc;
figure1=test1.plot_simulation('Ideal');
Elapsed time is 0.058935 seconds.
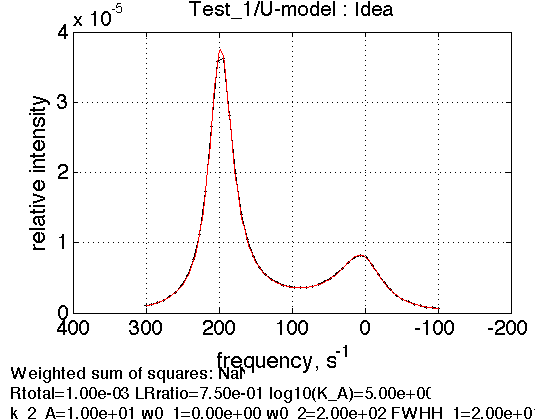
Adjust plotting ranges (if left empty - use automatic ranges in the plots)
test1.X_range=[min(test1.X) max(test1.X)]; test1.Y_range=[]; % re-plot figure1=test1.plot_simulation('Adjusted');
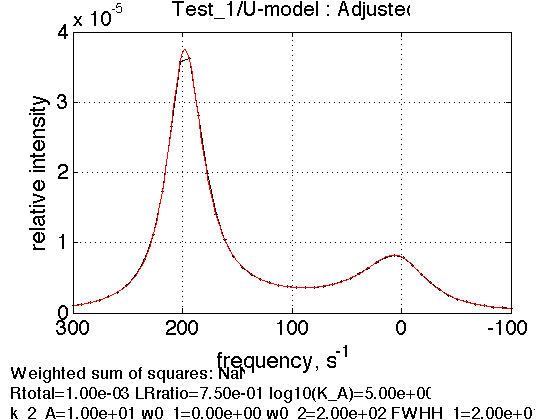
Simulate data with synthetic random noise
Copy dataset
test2=test1.make_a_copy('Test_2'); % Now add noise Signal_to_noise_ratio=20; Y_RMSD=max(test2.Y)/Signal_to_noise_ratio; test2.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure2=test2.plot_simulation('Noisy');

Save figure in all formats to a folder Tutorial_1/
results_output.output_figure(figure1,'Tutorial_1','U_model_ideal'); results_output.output_figure(figure2,'Tutorial_1','U_model_noisy');
Save a dataset object
We intend to do more work with this dataset later (try to fit it) therefore we need to save it to a disk. We will retrieve it in other tutorial using the following command temp=load('tutorial_1.mat'); test1=temp.test1; test2=temp.test2
save('tutorial_1.mat','test1', 'test2','setup');
Example of using numeric solution of the models
As example of using models with numeric solution I am showing U model switched to the numeric operation
options=optimset('Diagnostics','off', ... 'Display','off',... 'LargeScale','on',... 'LevenbergMarquardt', 'on',... 'TolFun',1e-18,... 'TolX',1e-18,... 'MaxFunEvals', 1e9); test1.set_active_model('U-model', 'lsqcurvefit', options) % select a model, solver and its options test1.show_active_model() tic; test1.simulate_noisy_data(parameters, 0, 0); toc; figure1=test1.plot_simulation('Ideal/numeric');
ans = Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: lsqcurvefit Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor Elapsed time is 0.372844 seconds.
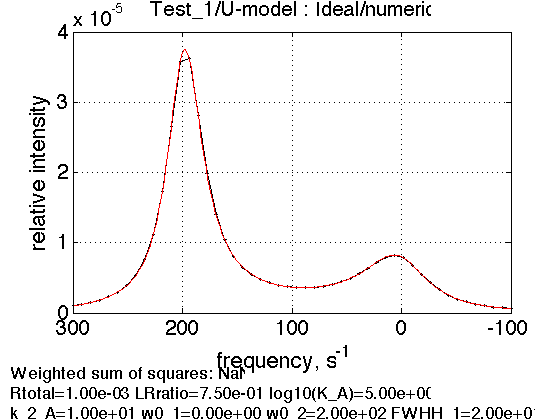
Estimation of the NMR line parameters
NOTE: Fitting with real models should be done in TotalFit session!
Here I show how to estimate parameters of the line to generate starting parameters for fitting with complex models: line position, line width.
In this spectrum we got two lines. We need: (1) trim the datasets to leave only one line, and (2) fit with Lorentzian function to obtain estimates of line geometry.
% copy dataset first (to be able to freely mess it up!) test3=test2.make_a_copy('parameter_estimation'); % trim data to retain only one line X1=100; X2=300; test3.trim_data(X1,X2); % show trimmed result test3.X_range=[X1 X2]; test3.plot_data('Trimmed');
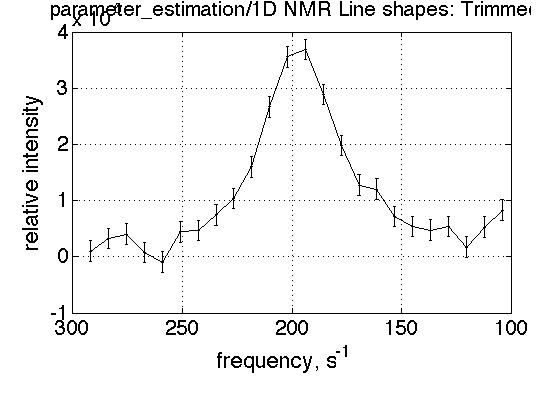
fit with lorentzian
mode='auto'; % use this if line is approximately centered in the window, % otherwise use 'manual' and provide initial parameters I0, FWHH and w0 % (intensity, Full-width-at-half-height and frequency). initial_parameters=[]; [ I0 FWHH w0 h1 h2] = test3.lorentzian_fit(mode, initial_parameters) % save figures results_output.output_figure(h1,'Tutorial_1','Lorentzian_initial_guess'); results_output.output_figure(h2,'Tutorial_1','Lorentzian_fit');
I0 =
7.5855e-04
FWHH =
40.2411
w0 =
196.2076
h1 =
6
h2 =
7


This procedure is very handy when fitting real experimental data by providing automatic initial guess.
Conclusions
Here we explored basic manipulation of the line shape dataset.