Tutorial 3. Simulate a 1D NMR titration series
by Evgenii Kovrigin, 05/17/2011
In this tutorial we will create a series of line shapes corresponding to an NMR titration experiment (gradual addition of a ligand to the receptor).
Contents
Clean up workspace (to avoid unwanted effects of old variable assignments)
close all clear all
CREATE A SERIES OF DATASETS FOR SIMULATIONS
Prepare a simulation session. Even though we are not planning to perform fitting of the data at this time, the simulation is better done using TotalFit object (session).
session_name='Tutorial_3_simulation';
session=TotalFit(session_name);
Create a simulation series
series_name='U_simulation'; % arbitrary dataset_class_name='NMRLineShapes1D'; % class name model_name='U-model'; model_numeric_solver='analytical'; model_numeric_options='no options'; number_of_datasets=5; number_of_points_to_simulate=100; X_min=-200; X_max=400; X_column=linspace(X_min, X_max, number_of_points_to_simulate)'; % use ' to transpose to a column-vector session.create_simulation_series(series_name, dataset_class_name, model_name, ... model_numeric_solver, model_numeric_options,... number_of_datasets, X_column);
Show datasets
%session.show_datasets % show all datasets in the session from all series session.show_series_datasets(series_name) % show only datasets of the series
ans = =========== Series "U_simulation" =============== ----------------------------- Dataset 1 --------------- Priority = 1 Name: U_simulation-1 Type: 1D NMR Line shapes Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor Dataset 2 --------------- Priority = 1 Name: U_simulation-2 Type: 1D NMR Line shapes Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor Dataset 3 --------------- Priority = 1 Name: U_simulation-3 Type: 1D NMR Line shapes Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor Dataset 4 --------------- Priority = 1 Name: U_simulation-4 Type: 1D NMR Line shapes Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor Dataset 5 --------------- Priority = 1 Name: U_simulation-5 Type: 1D NMR Line shapes Active model: Model 2: "U-model" Model description "1D NMR line shape for the U model, no temperature/field dependence, analytical" Model handle: line_shape_equations_1D.U_model_1D Current solver: analytical Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A) 4: k_2_A 5: w0_1 6: w0_2 7: FWHH_1 8: FWHH_2 9: ScaleFactor
ESTABLISH FITTING ENVIRONMENT
Prepare fitting environment. This is when we make 'session' object fully aware of presence of multiple datasets with (potentially) linkable parameters.
session.initialize_relation_matrix(sprintf('%s.all_datasets.txt', session_name))
Dataset listing is written to "Tutorial_3_simulation.all_datasets.txt".
To inspect current fitting environment use the following command:
session.generate_fitting_environment(sprintf('%s.fitting_environment.txt', session_name))
Fitting environment saved in Tutorial_3_simulation.fitting_environment.txt. ans = New parameter relations (indexed as in all_parameters vector): [1] 1:Rtotal [2] 1:LRratio [3] 1:log10(K_A) [4] 1:k_2_A [5] 1:w0_1 [6] 1:w0_2 [7] 1:FWHH_1 [8] 1:FWHH_2 [9] 1:ScaleFactor [10] 2:Rtotal [11] 2:LRratio [12] 2:log10(K_A) [13] 2:k_2_A [14] 2:w0_1 [15] 2:w0_2 [16] 2:FWHH_1 [17] 2:FWHH_2 [18] 2:ScaleFactor [19] 3:Rtotal [20] 3:LRratio [21] 3:log10(K_A) [22] 3:k_2_A [23] 3:w0_1 [24] 3:w0_2 [25] 3:FWHH_1 [26] 3:FWHH_2 [27] 3:ScaleFactor [28] 4:Rtotal [29] 4:LRratio [30] 4:log10(K_A) [31] 4:k_2_A [32] 4:w0_1 [33] 4:w0_2 [34] 4:FWHH_1 [35] 4:FWHH_2 [36] 4:ScaleFactor [37] 5:Rtotal [38] 5:LRratio [39] 5:log10(K_A) [40] 5:k_2_A [41] 5:w0_1 [42] 5:w0_2 [43] 5:FWHH_1 [44] 5:FWHH_2 [45] 5:ScaleFactor Summary of datasets with lookup vectors (position of each parameter in all_parameter vector) -------- Dataset 1/U_simulation-1, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 1 2 3 4 5 6 7 8 9 Dataset 2/U_simulation-2, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 10 11 12 13 14 15 16 17 18 Dataset 3/U_simulation-3, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 19 20 21 22 23 24 25 26 27 Dataset 4/U_simulation-4, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 28 29 30 31 32 33 34 35 36 Dataset 5/U_simulation-5, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 37 38 39 40 41 42 43 44 45
The all_parameters vector is the vector created of all parameters of all datasets. The parameters of each datasets are shown at their respective places in the all_parameters vector. If the parameters are linked---they will be shown together. Their respective 'individual' places are left unoccupied.
The lookup_vectors contain the same information in a reverse way: for each dataset they give positions of parameters as they are found in all_parameters vector. This is a 'reverse lookup', essentially.
CREATE PARAMETER LINKAGES
We have multiple datasets that are linked by their nature: they have many common parameters and just two parameters that is variable between them (the total receptor concentraion and ligand/receptor ratio). Enter here the parameter indices (as they show up in dataset listings) to link:
parameter_number_array=[ 3 4 5 6 7 8 9 ]; session.batch_link_parameters_in_series(series_name, parameter_number_array);
Propagate information about links across all datasets
session.generate_fitting_environment(sprintf('%s.fitting_environment.txt', session_name))
Fitting environment saved in Tutorial_3_simulation.fitting_environment.txt. ans = New parameter relations (indexed as in all_parameters vector): [1] 1:Rtotal [2] 1:LRratio [3] 1:log10(K_A) | 2:log10(K_A) | 3:log10(K_A) | 4:log10(K_A) | 5:log10(K_A) [4] 1:k_2_A | 2:k_2_A | 3:k_2_A | 4:k_2_A | 5:k_2_A [5] 1:w0_1 | 2:w0_1 | 3:w0_1 | 4:w0_1 | 5:w0_1 [6] 1:w0_2 | 2:w0_2 | 3:w0_2 | 4:w0_2 | 5:w0_2 [7] 1:FWHH_1 | 2:FWHH_1 | 3:FWHH_1 | 4:FWHH_1 | 5:FWHH_1 [8] 1:FWHH_2 | 2:FWHH_2 | 3:FWHH_2 | 4:FWHH_2 | 5:FWHH_2 [9] 1:ScaleFactor | 2:ScaleFactor | 3:ScaleFactor | 4:ScaleFactor | 5:ScaleFactor [10] 2:Rtotal [11] 2:LRratio [12] n/a [13] n/a [14] n/a [15] n/a [16] n/a [17] n/a [18] n/a [19] 3:Rtotal [20] 3:LRratio [21] n/a [22] n/a [23] n/a [24] n/a [25] n/a [26] n/a [27] n/a [28] 4:Rtotal [29] 4:LRratio [30] n/a [31] n/a [32] n/a [33] n/a [34] n/a [35] n/a [36] n/a [37] 5:Rtotal [38] 5:LRratio [39] n/a [40] n/a [41] n/a [42] n/a [43] n/a [44] n/a [45] n/a Summary of datasets with lookup vectors (position of each parameter in all_parameter vector) -------- Dataset 1/U_simulation-1, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 1 2 3 4 5 6 7 8 9 Dataset 2/U_simulation-2, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 10 11 3 4 5 6 7 8 9 Dataset 3/U_simulation-3, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 19 20 3 4 5 6 7 8 9 Dataset 4/U_simulation-4, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 28 29 3 4 5 6 7 8 9 Dataset 5/U_simulation-5, model "U-model" Parameters : Rtotal LRratio log10(K_A) k_2_A w0_1 w0_2 FWHH_1 FWHH_2 ScaleFactor Lookup vector : 37 38 3 4 5 6 7 8 9
As we requested, the first two parameters, Rtotal and LRratio, are individual for each dataset because they depend on how the sample was made. The rest of parameters belongs to the system (constants, frequencies, line widths) and to the hardware (ScaleFactor). They are made linked across entire series .
ASSIGN PARAMETERS
Choose some reasonable parameters (use ones from previous tutorials)
load('../Tutorial_1_2.U_simulation_fitting_one_dataset/tutorial_1.mat','setup');
To assign linked parameters we need to assign them to the first of the linked datasets (these values will be propagated to other datasets automatically through established links
Obtain number of the first datasets in the series
first_dataset_index=session.dataset_index(series_name,1);
prepare vectors for easier handling:
FAKE_VALUE=1E-6; % individual (unlinked) parameters: to be assigned later ideal_parameters=[ FAKE_VALUE FAKE_VALUE setup.Log10_K_A setup.k_2_A ... setup.w0_1 setup.w0_2 setup.FWHH_1 setup.FWHH_2 setup.ScaleFactor ] % no ';'-sign to show parameters
ideal_parameters =
Columns 1 through 5
0.0000 0.0000 5.0000 10.0000 0
Columns 6 through 9
200.0000 20.0000 20.0000 1.0000
Formally prepare limits (we will not need them for simulation but must assign arrays of the same size as parameters
fake_limits=zeros(1,length(ideal_parameters));
Perform assignment (I keep names of function arguments for information)
Monte_Carlo_range_min=fake_limits;
Monte_Carlo_range_max=fake_limits;
parameter_limits_min=fake_limits;
parameter_limits_max=fake_limits;
session.assign_parameter_values(first_dataset_index, ideal_parameters, ...
Monte_Carlo_range_min, Monte_Carlo_range_max, parameter_limits_min, parameter_limits_max);
Assign unlinked parameters These parameters are NOT linked therefore we need to go through all datasets to assign them.
Create a vector of values first
Rtotal_vector=ones(1,number_of_datasets)*setup.Rtotal; % this one is constant for all datasets LRratio_vector=[0 0.4 0.8 1.2 1.6]; % this one is variable
Create vector ranges for formal assignment (different length from the above!)
unlinked_fake_ranges=zeros(1,number_of_datasets);
Assign Rtotal
parameter_number=1; values_array=Rtotal_vector; MonteCarlo_range_min_array=unlinked_fake_ranges; MonteCarlo_range_max_array=unlinked_fake_ranges; parameter_limits_min_array=unlinked_fake_ranges; parameter_limits_max_array=unlinked_fake_ranges; session.assign_unlinked_parameter_in_series(... series_name, parameter_number, values_array, ... MonteCarlo_range_min_array, MonteCarlo_range_max_array, ... parameter_limits_min_array, parameter_limits_max_array);
Assign LRratio
parameter_number=2; values_array=LRratio_vector; session.assign_unlinked_parameter_in_series(... series_name, parameter_number, values_array, ... MonteCarlo_range_min_array, MonteCarlo_range_max_array, ... parameter_limits_min_array, parameter_limits_max_array);
CALCULATE MODEL VALUES
Run a simulation
X_standard_dev=0;
Y_standard_dev=1e-8; % something really small
session.simulate_series(series_name, X_standard_dev, Y_standard_dev);
Plot the simulation result
Y_offset_percent=10; % to offset figures vertically for easier viewing figure_handle1=session.plot_1D_series(series_name, 'Simulation 3', Y_offset_percent);

output simulation
figure_handle=figure_handle1; folder_name=session_name; figure_name=series_name; results_output.output_figure(figure_handle, folder_name, figure_name)
To adjust appearance of the output figures: load files from FIG/ folder in MATLAB and directly edit them there.
HOW TO CHECK VALUES OF SPECIFIC PARAMETERS FOR PARTICULAR DATASETS?
To do this we need (1) find out the index of the dataset we are interested in, and (2) display its 'best-fit' parameters (this is just the name of the vector simulated parameters are placed in);
The deal with the index is that we need TOTAL index the dataset has in the TotalFit object session. What is easy to know for us is the LOCAL index inside the series. This is how to convert the LOCAL index to a TOTAL one:
index_inside_the_series=3; dataset_in_question_index=session.dataset_index(series_name,index_inside_the_series);
Show parameters
session.dataset_best_fit_parameters(dataset_in_question_index)
ans =
Columns 1 through 5
0.0010 0.8000 5.0000 10.0000 0
Columns 6 through 9
200.0000 20.0000 20.0000 1.0000
Plot figure:
figure_title='inspection';
session.dataset_plot_fit(dataset_in_question_index, figure_title);
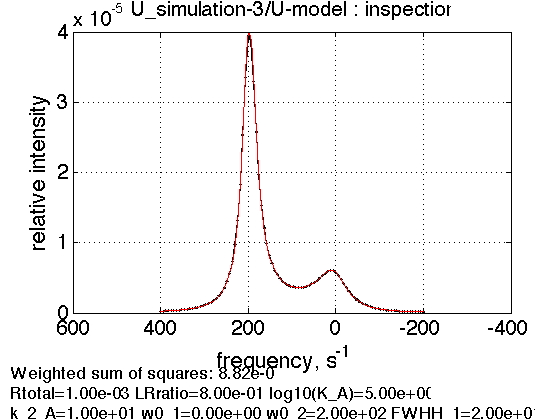
or (which uses index relative to the series)
session.plot_one_series_dataset(series_name,index_inside_the_series, figure_title);