B family models (analysis)
Binding of two ligand molecules to one receptor monomer including bidentate
ligand binding; macroscopic and microscopic consideration.
Evgenii L. Kovrigin
12-12-2014
Updates:
1/13/2015 - rename transitions to have internal isomerization ("hopping") transition called "B" in all models and bi-dentate binding transition called "C"
Name replacement schedule:
B_1_I_I -> C_I_I
B_1_I -> C_I
B_1 -> B
B_2 -> B
Here I am analyzing the numerical solutions for the B-family models
From: Equilibrium_thermodynamic_models/B_family_models/B
The more complicated version where the ligand is bidentate and capable of binding at both sites:
From Equilibrium_thermodynamic_models/B_family_models/B-bidentateL
This may be viewed as a combination of B and U-RL model (not exactly, though, because there are three isomerization transitions in B-bidentateL-micro.
clean up workspace
reset()
STEP 1: Set path to saved derivation results
NOTE: make sure the path ends with slash character "/".
ProjectName:="B_family_models_derivation";
CurrentPath:="/Volumes/Leopard_Partition/Users/kovrigin/Documents/Workspace/Global Analysis/IDAP/Mathematical_models/Equilibrium_thermodynamic_models/B_family_models/";
![]()
![]()
Read results of derivations
filename:=CurrentPath.ProjectName.".mb";
fread(filename,Quiet):
anames(User);


STEP 2: Assume values and test procedures:
Rtot_value:=1e-3:
K_A_1_value:= 1e7:
K_A_2_value:= 1e8:
LR_ratio_value:= 0.8:
K_A_1_I_value:=2e4:
K_A_1_I_I_value:=1e2:
K_A_2_I_I_value:=3e5:
K_C_I_I_value:=11:
Test calculations:
pnLeq_BmAcro(Rtot_value, LR_ratio_value, K_A_1_value, K_A_2_value);
pnReq_BmAcro(Rtot_value, LR_ratio_value, K_A_1_value, K_A_2_value);
pnRLeq_BmAcro(Rtot_value, LR_ratio_value, K_A_1_value, K_A_2_value);
pnRL2eq_BmAcro(Rtot_value, LR_ratio_value, K_A_1_value, K_A_2_value);
![]()
![]()
![]()
![]()
pnLeq_Bmicro(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value);
pnReq_Bmicro(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value);
pnRLIIeq_Bmicro(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value);
pnRLIeq_Bmicro(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value);
pnRL2eq_Bmicro(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value);
![]()
![]()
![]()
![]()
![]()
pnLeq_Bbidentate(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value);
pnReq_Bbidentate(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value);
pnRLIeq_Bbidentate(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value);
pnRLIIeq_Bbidentate(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value);
pnRLbieq_Bbidentate(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value);
pnRL2eq_Bbidentate(Rtot_value, LR_ratio_value, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value);
![]()
![]()
![]()
![]()
![]()
![]()
=> all operational
STEP 3: Make wrapper functions for plotting dependent only on one parameter x=L/R:
fnLeq_BmAcro:=x -> pnLeq_BmAcro(Rtot_value, x, K_A_1_value, K_A_2_value):
fnReq_BmAcro:=x -> pnReq_BmAcro(Rtot_value, x, K_A_1_value, K_A_2_value):
fnRLeq_BmAcro:=x -> pnRLeq_BmAcro(Rtot_value, x, K_A_1_value, K_A_2_value):
fnRL2eq_BmAcro:=x -> pnRL2eq_BmAcro(Rtot_value, x, K_A_1_value, K_A_2_value):
fnLeq_Bmicro:=x -> pnLeq_Bmicro(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value):
fnReq_Bmicro:=x -> pnReq_Bmicro(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value):
fnRLIIeq_Bmicro:=x -> pnRLIIeq_Bmicro(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value):
fnRLIeq_Bmicro:=x -> pnRLIeq_Bmicro(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value):
fnRL2eq_Bmicro:=x -> pnRL2eq_Bmicro(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value):
fnLeq_Bbidentate:=x -> pnLeq_Bbidentate(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value):
fnReq_Bbidentate:=x -> pnReq_Bbidentate(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value):
fnRLIeq_Bbidentate:=x -> pnRLIeq_Bbidentate(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value):
fnRLIIeq_Bbidentate:=x -> pnRLIIeq_Bbidentate(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value):
fnRLbieq_Bbidentate:=x -> pnRLbieq_Bbidentate(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value):
fnRL2eq_Bbidentate:=x -> pnRL2eq_Bbidentate(Rtot_value, x, K_A_1_I_value, K_A_1_I_I_value, K_A_2_I_I_value, K_C_I_I_value):
2.1 Macroscopic B model
ModelName:="B macro":
// Thermodynamic parameters
Rtot_value:=1e-3:
K_A_1_value:= 1e4:
K_A_2_value:= 1e4:
LR_ratio_max:= 6:
// Plotting parameters
LW:=1: x_min:=1e-6:
// Compute dependent constants
// None in this model
p_Leq_BmAcro:= plot::Function2d(Function=(fnLeq_BmAcro), LegendText="[L]", Color = RGB::Green, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_Req_BmAcro:= plot::Function2d(Function=(fnReq_BmAcro), LegendText="[R]", Color = RGB::Black, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RLeq_BmAcro:= plot::Function2d(Function=(fnRLeq_BmAcro), LegendText="[RL]", Color = RGB::Blue, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RL2eq_BmAcro:= plot::Function2d(Function=(fnRL2eq_BmAcro), LegendText="[RL2]", Color = RGB::Red, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
// Report constants
print(Unquoted,"Model: ".ModelName);
print(Unquoted,"R(total):=".Rtot_value.":");
print(Unquoted,"K_A_1:=".K_A_1_value.":");
print(Unquoted,"K_A_2:=".K_A_2_value.":");
// Plot all
// plot all together
plot(p_Leq_BmAcro,p_Req_BmAcro, p_RLeq_BmAcro,p_RL2eq_BmAcro, YAxisTitle="M", Header=("Model: ".ProjectName), Height=160, Width=100,TicksLabelFont=["Helvetica",12,[0,0,0],Left], AxesTitleFont=["Helvetica",14,[0,0,0],Left], XGridVisible=TRUE, YGridVisible=TRUE,LegendVisible=TRUE, LegendFont=["Helvetica",14,[0,0,0],Left]);
Model: B macro
R(total):=0.001:
K_A_1:=10000.0:
K_A_2:=10000.0: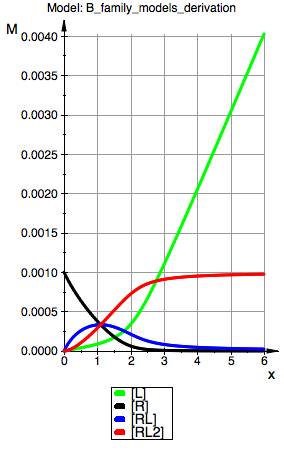
Summary
|
Remove second binding event |
Equal microscopic binding constants (identical sites, no cooperativity between sites); K1=4*K2 |
Equal mAcroscopic binding constants (identical sites, positive cooperativity) |
|
|
|
Model: B macro |
Model: B macro |
Model: B macro |
|
|
|
|
|
|
|
|
=> Model behaves as expected
2.2 Microscopic B model (mono-dentate binding only
ModelName:="B micro":
// Thermodynamic parameters
Rtot_value:=1e-3:
K_A_1_I_value:=5000:
K_A_1_I_I_value:=5000:
K_A_2_I_I_value:=0:
LR_ratio_max:= 3:
// Plotting parameters
LW:=1: x_min:=1e-6:
// Compute dependent constants
K_A_2_I_value:= eqKA2I_Bmicro[2] | K_A_1_I=K_A_1_I_value | K_A_1_I_I=K_A_1_I_I_value | K_A_2_I_I=K_A_2_I_I_value:
K_B_value:= eqKB1_Bmicro[2] | K_A_1_I=K_A_1_I_value | K_A_1_I_I=K_A_1_I_I_value | K_A_2_I_I=K_A_2_I_I_value:
p_Leq_Bmicro:= plot::Function2d(Function=(fnLeq_Bmicro), LegendText="[L]", Color = RGB::Green, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_Req_Bmicro:= plot::Function2d(Function=(fnReq_Bmicro), LegendText="[R]", Color = RGB::Black, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RLIeq_Bmicro:= plot::Function2d(Function=(fnRLIeq_Bmicro), LegendText="[RLI]", Color = RGB::Blue, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RLIIeq_Bmicro:= plot::Function2d(Function=(fnRLIIeq_Bmicro), LegendText="[RLII]", Color = RGB::Cyan, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RL2eq_Bmicro:= plot::Function2d(Function=(fnRL2eq_Bmicro), LegendText="[RL2]", Color = RGB::Red, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
// Report constants
print(Unquoted,"Model: ".ModelName);
print(Unquoted,"R(total):=".Rtot_value.":");
print(Unquoted,"K_A_1_I:=".K_A_1_I_value.":");
print(Unquoted,"K_A_1_I_I:=".K_A_1_I_I_value.":");
print(Unquoted,"K_A_2_I_I:=".K_A_2_I_I_value.":");
print(Unquoted,"K_A_2_I:=".K_A_2_I_value." (dep):");
print(Unquoted,"K_B:=".K_B_value." (dep):");
// Plot all
// plot all together
plot(p_Leq_Bmicro,p_Req_Bmicro, p_RLIeq_Bmicro,p_RLIIeq_Bmicro,p_RL2eq_Bmicro, YAxisTitle="M", Header=("Model: ".ProjectName), Height=160, Width=100,TicksLabelFont=["Helvetica",12,[0,0,0],Left], AxesTitleFont=["Helvetica",14,[0,0,0],Left], XGridVisible=TRUE, YGridVisible=TRUE,LegendVisible=TRUE, LegendFont=["Helvetica",14,[0,0,0],Left]);
Model: B micro
R(total):=0.001:
K_A_1_I:=5000:
K_A_1_I_I:=5000:
K_A_2_I_I:=0:
K_A_2_I:=0 (dep):
K_B:=1 (dep):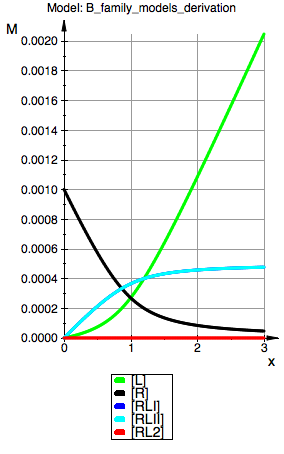
Summary of test cases
|
Turn off the second transition (identical sites) |
No cooperativity between two identical sites: K1=2K, K2=K/2 (matching B macro simulation) |
Positive cooperativity, identical sites (matching B-macro simulation) |
No cooperativity, different affinity of two sites |
Positive cooperativity, different affinity between two sites |
|
|
|
|
|
|
Model: B micro R(total):=0.001: |
Model: B micro |
Model: B micro |
Model: B micro |
Model: B micro |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
=> Model behaves as expected: reproduces behavior of the B macro if sites are set to be identical. Offers microscopic view as advertised.
2.3 Microscopic B-bidentateL model (bi-dentate ligand)
eqKB2_Bbidentate
![]()
ModelName:="B-bidentateL":
// Thermodynamic parameters
Rtot_value:=1e-3:
K_A_1_I_value:=5000:
K_A_1_I_I_value:=2500:
K_A_2_I_I_value:=20000:
K_C_I_I_value:=10:
LR_ratio_max:= 3:
// Plotting parameters
LW:=1: x_min:=1e-6:
// Compute dependent constants
K_A_2_I_value:= eqKA2I_Bbidentate[2] | K_A_1_I=K_A_1_I_value | K_A_1_I_I=K_A_1_I_I_value | K_A_2_I_I=K_A_2_I_I_value | K_C_I_I=K_C_I_I_value:
K_C_I_value:= eqKB1I_Bbidentate[2] | K_A_1_I=K_A_1_I_value | K_A_1_I_I=K_A_1_I_I_value | K_A_2_I_I=K_A_2_I_I_value | K_C_I_I=K_C_I_I_value:
K_B_value:= eqKB2_Bbidentate[2] | K_A_1_I=K_A_1_I_value | K_A_1_I_I=K_A_1_I_I_value | K_A_2_I_I=K_A_2_I_I_value | K_C_I_I=K_C_I_I_value:
p_Leq_Bbidentate:= plot::Function2d(Function=(fnLeq_Bbidentate), LegendText="[L]", Color = RGB::Green, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_Req_Bbidentate:= plot::Function2d(Function=(fnReq_Bbidentate), LegendText="[R]", Color = RGB::Black, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RLIeq_Bbidentate:= plot::Function2d(Function=(fnRLIeq_Bbidentate), LegendText="[RLI]", Color = RGB::Blue, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RLIIeq_Bbidentate:= plot::Function2d(Function=(fnRLIIeq_Bbidentate), LegendText="[RLII]", Color = RGB::Cyan, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RL2eq_Bbidentate:= plot::Function2d(Function=(fnRL2eq_Bbidentate), LegendText="[RL2]", Color = RGB::Red, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
p_RLbieq_Bbidentate:= plot::Function2d(Function=(fnRLbieq_Bbidentate), LegendText="[RLbi]", Color = RGB::Magenta, XMin=(x_min),XMax=(LR_ratio_max),XName=(x),TitlePositionX=(0) ,LineWidth=LW):
// Report constants
print(Unquoted,"Model: ".ModelName);
print(Unquoted,"R(total) = ".Rtot_value);
print(Unquoted,"K_A_1_I = ".K_A_1_I_value);
print(Unquoted,"K_A_2_I = ".K_A_2_I_value." (dep)");
print(Unquoted,"K_A_1_I_I = ".K_A_1_I_I_value);
print(Unquoted,"K_A_2_I_I = ".K_A_2_I_I_value);
print(Unquoted,"K_C_I = ".K_C_I_value." (dep)");
print(Unquoted,"K_C_I_I = ".K_C_I_I_value);
print(Unquoted,"K_B = ".K_B_value." (dep)");
// Plot all
// plot all together
plot(p_Leq_Bbidentate,p_Req_Bbidentate, p_RLIeq_Bbidentate,p_RLIIeq_Bbidentate,p_RLbieq_Bbidentate,p_RL2eq_Bbidentate, YAxisTitle="M", Header=("Model: ".ProjectName), Height=160, Width=100,TicksLabelFont=["Helvetica",12,[0,0,0],Left], AxesTitleFont=["Helvetica",14,[0,0,0],Left], XGridVisible=TRUE, YGridVisible=TRUE,LegendVisible=TRUE, LegendFont=["Helvetica",14,[0,0,0],Left]);
Model: B-bidentateL
R(total) = 0.001
K_A_1_I = 5000
K_A_2_I = 40000 (dep)
K_A_1_I_I = 2500
K_A_2_I_I = 20000
K_C_I = 20 (dep)
K_C_I_I = 10
K_B = 1/2 (dep)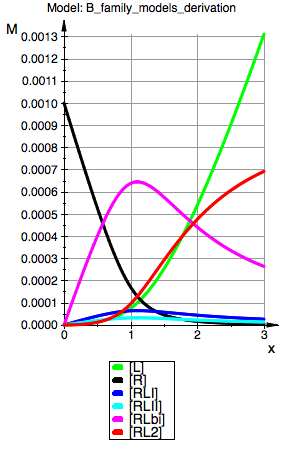
Summary of test cases
First, reduce this model to previous one: no bidentate binding
|
no second binding step (matching B micro case above) |
No cooperativity between two identical sites: K1=2K, K2=K/2 (matching B macro and B micro simulation above) |
Positive cooperativity, identical sites (matching B-micro simulation) |
No cooperativity, different affinity of two sites |
Positive cooperativity, different affinity between two sites |
|
Model: B-bidentateL |
Model: B-bidentateL |
Model: B-bidentateL |
Model: B-bidentateL |
Model: B-bidentateL |
|
|
|
|
|
|
|
Include bi-dentate binding mode |
Include bi-dentate binding mode |
Include bi-dentate binding mode |
Include bi-dentate binding mode |
Include bi-dentate binding mode |
|
Model: B-bidentateL |
Model: B-bidentateL |
Model: B-bidentateL |
Model: B-bidentateL |
Model: B-bidentateL |
=> This model completely replicates behavior of B micro if we turn off bidentate binding.
=> Addition of bi-dentate binding produces sensible result
Conclusions
All three B models work, reproduce corresponding simpler ones and are ready for use in IDAP.