
R2
Derivation of differential equations describing evolution of spin populations

This document accompanies Lecture 5, last section.
2. Microscopic reversibility law
4. Expression in terms of spin (monomer) concentrations
clean up workspace
reset()
Write properly balanced reactions equations:
(1) (2)
2R <=> R2
Constants: k_12, k_21.
Write reaction rates
We distinguish reaction rates (elementary reaction acts per unit time) and conversion rates (number of moles of specific species consumed/produced per unit time). Conversion rates, dc/dt, for species are related to reaction rates, Rate, through molecularity coefficients.
Dimerization rates
a reaction rate:
eq1_1:= Rate_1_2 = k_1_2*R*R
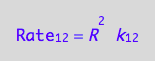
a conversion rate:
eq1_2a:= dcRdt_1_2 = Rate_1_2*2
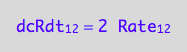
-> this reflects the fact that two molecules of R are consumed to make R2 dimer.
The final form:
eq1_2b:= eq1_2a | eq1_1
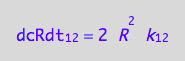
Dissociation rates
a reaction rate:
eq1_3:= Rate_2_1 = k_2_1*R2
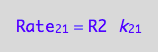
a conversion rate:
eq1_4a:= dcR2dt_2_1 = Rate_2_1
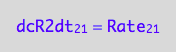
The final form:
eq1_4b:= eq1_4a | eq1_3
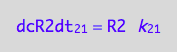
The principle of microscopic reversebility requires that, in equilibrium, forward reaction rate is equal to reverse reaction rate:
eq2_1:= Rate_1_2 = Rate_2_1;
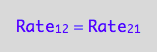
Express in conversion rates terms:
solve(eq1_2a, Rate_1_2):
%[1]:
eq2_2:= Rate_1_2 = %
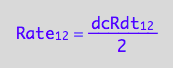
solve(eq1_4a, Rate_2_1):
%[1]:
eq2_3:= Rate_2_1 = %
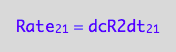
eq2_4:= eq2_1 | eq2_2 | eq2_3
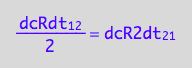
-> this result again reflects the fact that two molecules of R make the R2 dimer.
To define evolution of the species we need to compute concentrations of a monomer and a dimer as a function of time. To this end, we will write differential equations for conversion rates of the species.
Net conversion rate of the monomer R
In a reversible process both forward and reverse reaction occur simultaneously. Thus, the net conversion rate of R is a difference between the conversion rate of R in the association and in dissociation reactions:
eq3_1:= dcRdt_N= - dcRdt_1_2 + dcRdt_2_1

The dimerization term (12) is known
eq1_2b
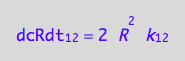
We do not have d[R]/dt of the dissociation reaction (21 term). However, we know that the mass conservation law requires that every molecule of R2 makes two molecules of R. Therefore:
eq3_3:= dcRdt_2_1 = 2*dcR2dt_2_1
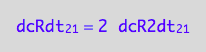
which is
eq3_4:= eq3_3 | eq1_4b
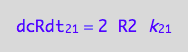
Substitute to the equation for net conversion rates:
eq3_1;
% | eq3_4:
% | eq1_2b :
eq3_6:= %


=> THIS IS OVERALL CONVERSION RATE OF R SPECIES
Overall conversion rate of the dimer, R2
Similarly to the above, the net conversion rate of the dimer is:
eq3_7:= dcR2dt_N = -dcR2dt_2_1 + dcR2dt_1_2

The rate for dissociation is known
eq1_4b
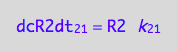
We do not have d[R2]/dt of the dimerizaion reaction (12 term). However, we know that the mass conservation law requires that every molecule of R2 needs two molecules of R. Therefore:
eq3_8:= dcR2dt_1_2 = dcRdt_1_2/2
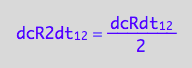
which is
eq3_9:= eq3_8 | eq1_2b

Now, substitute in the equation for a net rate
eq3_7;
% | eq1_4b | eq3_9:
eq3_10:= %

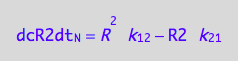
=> THIS IS OVERALL CONVERSION RATE OF R2 SPECIES
The Bloch-McConnell equations describe evolution of bulk magnetization of a sample, which is proportional to the number of spins found in every specific magnetic environment. A dimer contains two identical spins, therefore the amount of magnetization from spins in the environment of dimer is proportional to the doubled equilibrium concentration of a dimer.
Define new variables for concentrations and net conversion rates
A monomeric species contains only one spin so everything is equivalent:
eq4_1:= C_1 = R;
eq4_2:= dC_1dt=dcRdt_N;
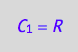

Dimeric species contains two spins:
eq4_3:= C_2 = 2*R2;
eq4_4:= dC_2dt=2*dcR2dt_N
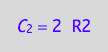
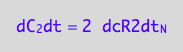
Express monomer/dimer concentration in terms of spin concentrations for easier substitution
solve(eq4_1,R):
eq4_5:= R=%[1]
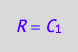
solve(eq4_3, R2):
eq4_6:= R2 = %[1]
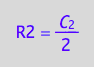
Substitute in net conversion rates
A spin in a monomer
eq4_2 | eq3_6;
% | eq4_5 | eq4_6;
eq4_7:= %:

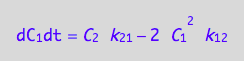
A spin in a dimer
eq4_4 | eq3_10;
% | eq4_5 | eq4_6;
eq4_8:= %:
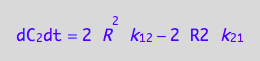
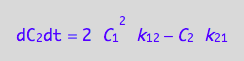
Summarize the derivation results
These are the differential equations governing concentrations of spins experiencing environment of a monomer, C1, and a dimer, C2.
eq4_7;
eq4_8;
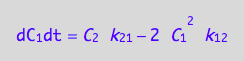
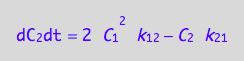
where
eq4_1;
eq4_3;
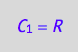
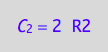
Prepare results for transfer to MATLAB
To avoid typing errors when transfering derived K matrix to MATLAB we enter it here and then directly test against derivation result. Then K matrix may be transfered to MATLAB by cut-and-paste of the MuPad output.
Restate the results
eq4_7;
eq4_8;
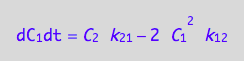
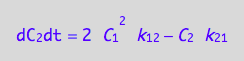
Enter the K-matrix looking at the above results.
Simple rules that allow catching mistakes in K matrix derivation:
(1) a sum of each column should be zero (so each constant must appear with both positive and negative sign), and
(2) each row has to have complete pairs of constants (i.e., if k12
appears there must be k21 in the same row with an opposite sign and so on).
K:=matrix(2,2,[
[ -2*k_1_2*C_1 , k_2_1 ],
[ 2*k_1_2*C_1, -k_2_1 ]
])
![matrix([[-2*C_1*k_1_2, k_2_1], [2*C_1*k_1_2, -k_2_1]])](R2_images/math38.png)
Create P-column vector
P:=matrix(2,1,[ C_1, C_2])
![matrix([[C_1], [C_2]])](R2_images/math39.png)
Check correctness of the entered K matrix by multiplying with P and comparing to the above equations:
Multiply K and P:
dCdt_manual_input:=K*P
![matrix([[C_2*k_2_1 - 2*C_1^2*k_1_2], [2*C_1^2*k_1_2 - C_2*k_2_1]])](R2_images/math40.png)
Collect right-hand-side parts of equations
dCdt_mupad:=matrix(2,1,[ rhs(eq4_7), rhs(eq4_8) ])
![matrix([[C_2*k_2_1 - 2*C_1^2*k_1_2], [2*C_1^2*k_1_2 - C_2*k_2_1]])](R2_images/math41.png)
Compare derivation result to manual input
dCdt_mupad=dCdt_manual_input:
normal(%);
bool(%)
![matrix([[C_2*k_2_1 - 2*C_1^2*k_1_2], [2*C_1^2*k_1_2 - C_2*k_2_1]]) = matrix([[C_2*k_2_1 - 2*C_1^2*k_1_2], [2*C_1^2*k_1_2 - C_2*k_2_1]])](R2_images/math42.png)

=> Great!
Use this K-matrix (copy-paste output to MATLAB)
K;
![matrix([[-2*C_1*k_1_2, k_2_1], [2*C_1*k_1_2, -k_2_1]])](R2_images/math44.png)
Development of differential equations governing spin populations in systems with oligomerization is demonstrated.