

In this document I follow my introduction of three state models for fitting of series of 1D NMR datasets in IDAP. I will simulate the line shape data with LineShapeKin Simulation and then demonstrate how line shape fitting may be done with IDAP. The user may follow the same protocol to fit real experimental data with these example workflows.
Fast exchange scenario
Slow exchange scenario
| Expected parameters | Initial approximation: |
log10(K_A1)= 4 log10(K_A2) = 4 k_2_A1= 1000 k_2_A2= 10 w0_RL= 200 w0_RL2 = 300 FWHH_RL= 10 FWHH_RL2 = 10 |
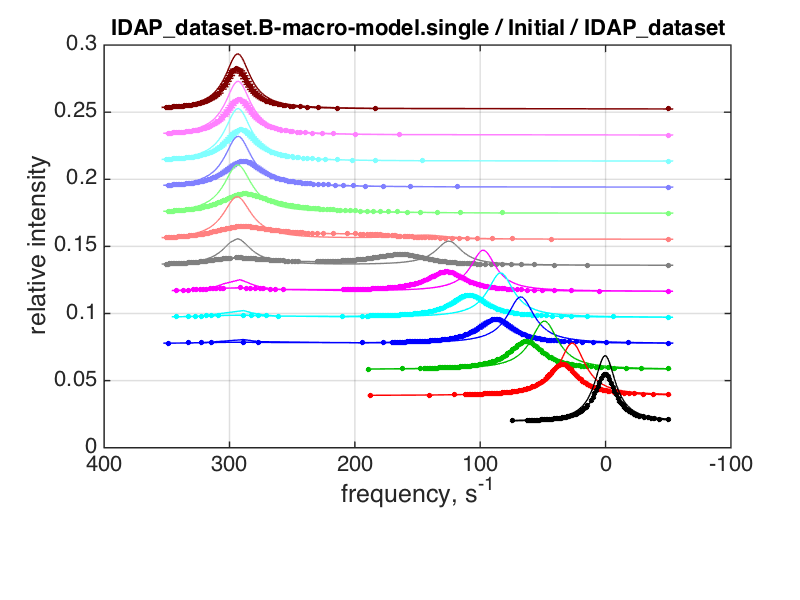 |
| GlobalSearch | Simplex | |
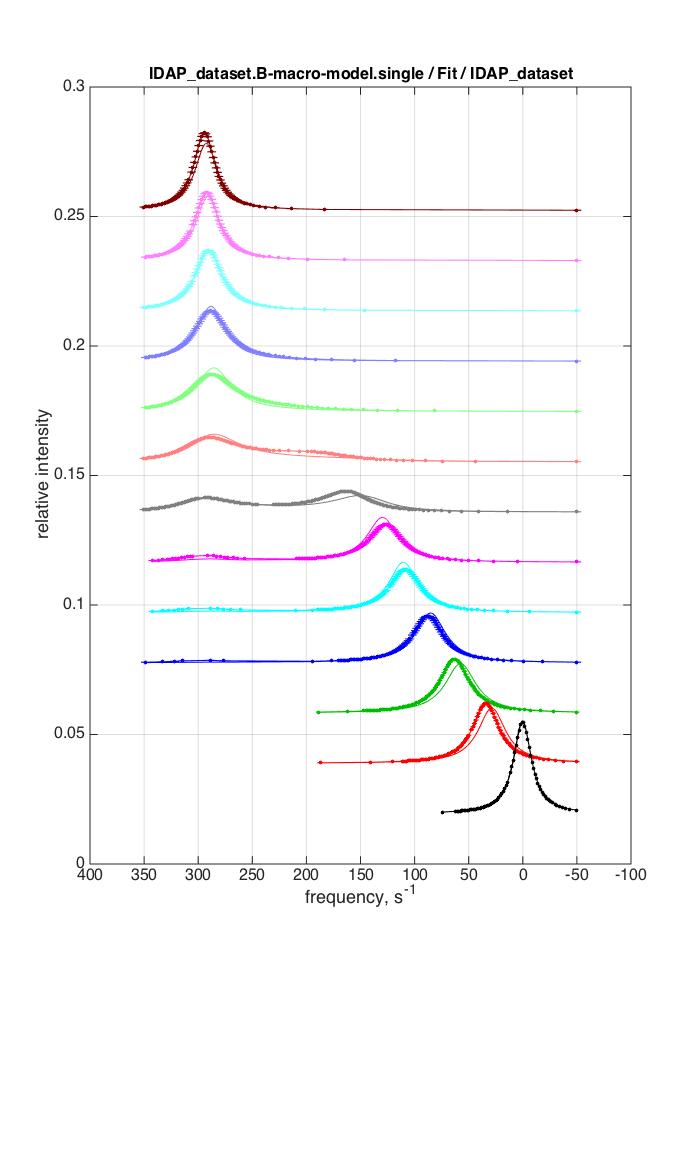 |
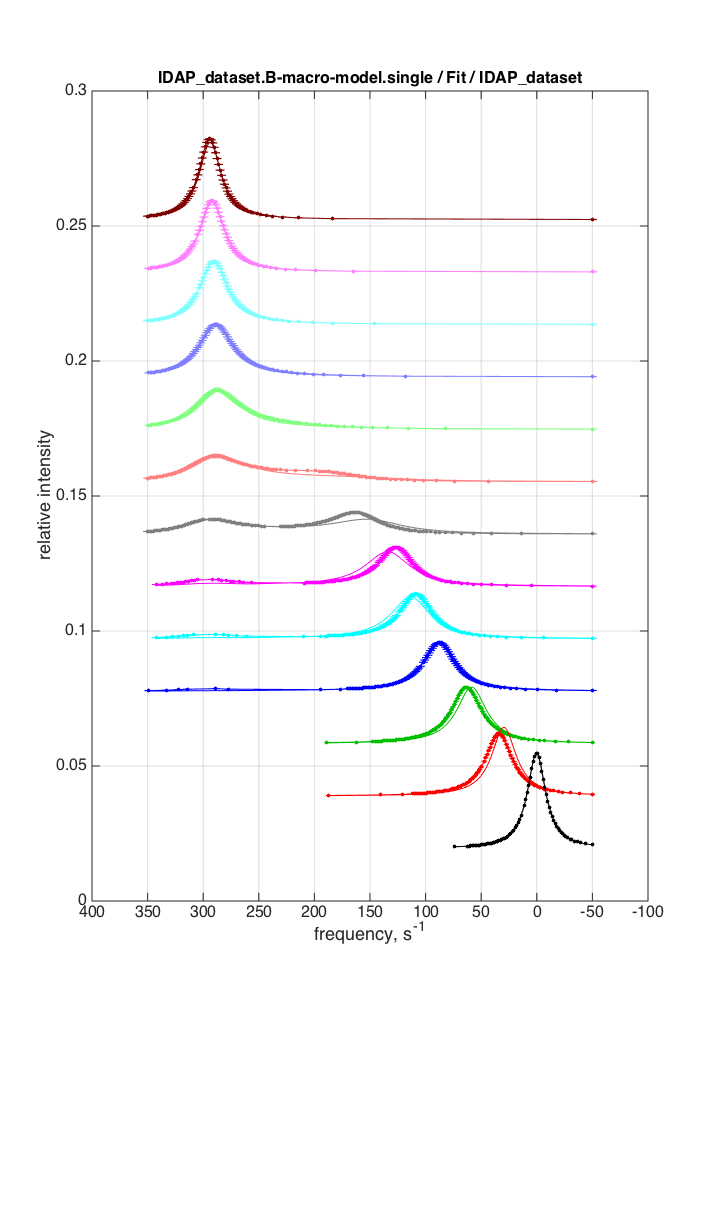 |
|
|
log10(K_A1) = 5.58 log10(K_A2)= 4.46 k_2_A1= 6.227387e+02 :k_2_A2 = 9.465268e+00 w0_RL = 1.479257e+02 ( w0_RL2 = 2.956745e+02 FWHH_RL = 3.011751e+01 FWHH_RL2 = 2.425213e+01 ScaleFactor = 3.286271e+02 Total sum of squares (SS) = 1.15811 Worse than simplex |
log10(K_A1)= 5.6 log10(K_A2) = 4.2 k_2_A1= 5.436067e+03 k_2_A2= 1.033124e+01 w0_RL= 1.486588e+02 w0_RL2 = 2.985160e+02 FWHH_RL= 4.800909e+01 FWHH_RL2 = 2.054429e+01 ScaleFactor= 3.409475e+02 Total sum of squares (SS) = 0.889889 |
|
| Fitting time: 1100 sec = 20 min | Fitting time: 200 sec = 3 min |
CONCLUSIONS:
Done in Tutorial 6: tutorial_6_testing_new_model
IDAP/IDAP_Online_Documentation/Tutorials/NMR_line_shapes/Tutorial_8.Overview_and_testing_three_state_model/U_R2_FWHHconstr_model
Compare to simulations with LineShapeKin Simulation
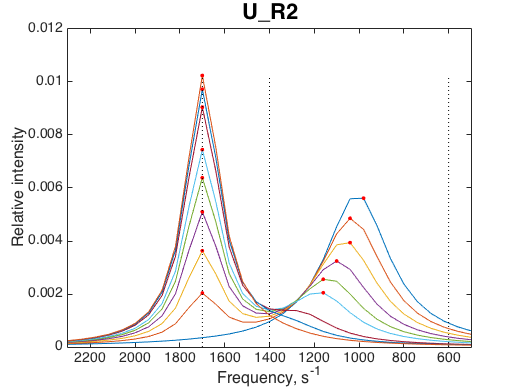 |
# Unique model identifier # Model description # Association constants # Rate constants of REVERSE reactions # Names of NMR-active species # Names of NMR unobservable species # Chemical shifts of pure species, 1/s # Relaxation rates of pure species, 1/s # Heat of formation of the species, relative units
|
Observation: The IDAP model code reproduced the LineShapeKin result.
test_fitting/
main_U_R2_FWHHconstr_model_1D.m
INITIAL
|
l |
SIMPLEX Fitting time: 41 sec
|
|
|
log10(K_A)= 4.1e+00 log10(K_B)= 3.2e+00 k_2_A = 5.7 k_2_B = 3.3e+03 w0_R = 1.4e+03 w0_R2 = 7.110175e+02 FWHH_R_monomer = FWHH_RL = 1.896252e+02 ScaleFactor = 3.524467e+02 Total sum of squares (SS) = 0.00418529 |
|
Conclusions:
Implementation of the U_R2_FWHHconstr-model in IDAP was validated