

NOTE: Prior to use, KovriginNMR code should be placed in appropriate locations by the NMR manager as described in Installation and Administration.
It is critical for KovriginNMR operation that Topspin Spooler does not interfere with control of a spectrometer by automatically intercepting user's commands. To disable automatic spooling, navigate into Manage:
choose Preferences:
and verify that "Enable automatic command spooling" is unchecked:
Click Apply and Close to save the setting and exit the dialog
NOTE: If the checkbox remains checked, you may see error messages during operation of KovriginNMR that resemble hardware failures because Spooler will be trying to control the spectrometer at the same time with KovriginNMR.
3. Choose User-Defined Buttons:
4. The dialog opens with three columns: Name or Icon, Command, and Tool Tip Text:
NOTE: A popup window with dimensions (Step 8) below may be displayed before this one. Follow Step 8 instructions then.
5. Type in the following:
Name: KovriginNMR,
Command column: knmr_general_launch_fbar,
Tool Tip Text: NMR Workflows
6. Make sure the checkbox Disable display of user-defined buttons on the bottom of the dialog is unchecked.
7. Click OK to close the dialog.8. A window asking to select a dimension pops up. Choose 1D-nD:
9. You should see a new button titled KovriginNMR appearing on the toolbar:
If Topspin glitches and it does not happen, you should quit, re-launch Topspin, and open any dataset to see the KovriginNMR button.
IMPORTANT: The toolbar button will only appear when some NMR dataset is open!
10. Sometimes, Topspin still glitches and the KovriginNMR button remains invisible. In this case, click on Less icons button
this mode folds the toobar to one row and displays the KovriginNMR button:
Generally, exiting Topspin, logging out and back in, and re-launching Topspin should rectify all problems.
Activate KovriginNMR toolbar
Clicking on KovriginNMR button will bring up a set of new buttons in the flowbar:
IMPORTANT: When you click the KovriginNMR button, it replaces the current flowbar with its own buttons. By design of Topspin, the current tab (Acquire) in the above image from this point on holds these buttons and loses its own. To bring the original Acquire buttons back, please, click Back to Acquire.
Overview of buttons
Return to Acquire - reinstates the Acquire tab buttons
New Topspin - opens an additional Topspin window
Open Workflow - allows access to the Python folder of Topspin and to the KovriginNMR template script
Stop Workflow - allows to halt a running KovriginNMR script
Show Most Recent Report - launches a web browser with the last HTML report created by KovriginNMR operation.
Create a new KovriginNMR workflow (script)
EK>>>>> UPDATE THIS SECTION WITH NEW PICTURES!!!!
To create a new script using KovriginNMR libraries and commands, click on Open Workflow and select Open KovriginNMR workflow template:
This brings up a Python editor with a template file, you can use to create your own KovriginNMR scripts:
This is a template common for all KovriginNMR users and serves as a source of examples of code you may want to use.
To make your own script, the first action is to re-save the file with a new name. Navigate to File
and click on Save as to bring up the Save as... dialog. In this dialog, first, you have to make sure that the Destination folder is /user
if it is not, click on the arrow down symbol and select it :
Next, you enter your new file name.
IMPORTANT: Topspin holds Python files of all users in the same folder. This creates a certain difficulty in searching for your own scripts if other users also actively make their own. Therefore, there are two rules to follow when choosing a file name:
Rule 1: Start the new name of your workflow with your initials and an underscore symbol. This will make your workflows appear together in the file browser and will make it easier to distinguish them from files other users.
Rule 2: Finish your file name with '.py' to indicate to Topspin that this is a Python program.
For example, my test workflow will be ek_test1.py
After you re-saved the template with your new file name, you own the file and can edit it. The KovriginNMR template has been written as a readable document showcasing some possibilites you may find useful. For a systematic description of the KovriginNMR classes and a code, please, see Python classes and modules. The template header contains several mandatory commands to enable KovriginNMR operation. These commands are followed by multiple optional sections, which are all commended out and, therefore, ignored by Python interpreter. You can uncomment a desired section and delete the rest of commented text. Please, remember that you can always bring back the original template through Open Workflow: Open KovriginNMR workflow template and start from scratch if needed.
Take the following steps:
- Read the instructions in the header of the file
- Find the section resembling what you need to do
- Uncomment this section by removing '#' at the beginning of the lines.
- Delete the remaining commented lines of other sections to make your script shorter.
- Save the script through File:Save in the menu bar
- Further edit the workflow as needed for your project.
- Save the final version of the script.
Once you created your own script, it is saved in /opt/topspin/exp/stan/nmr/py/user on the NMR workstation hard drive. You may reopen it by choosing Open Workflow: Open Python Browser:
This action brings up a File Open dialog. Please, check what the Source folder is. By default it will open in a system Python folder:
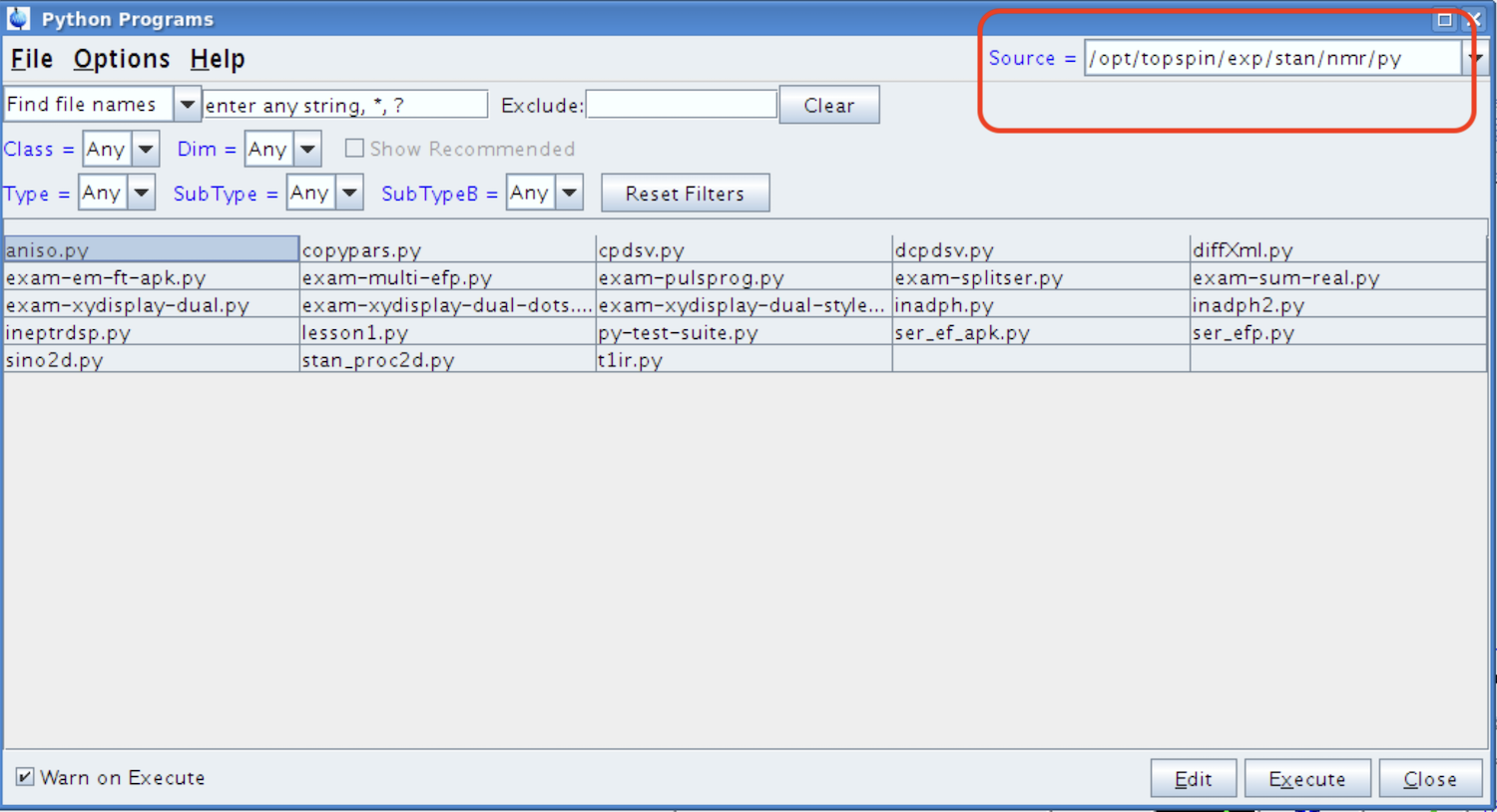
If it is not py/user, you need to change the path:
Finally, you arrive to the folder where your file was saved. Find it in a list, highlight, and click Edit to open your file:
NOTE: Prior to launching KovriginNMR script, first, you must display some (any) experiment in Topspin. The KovriginNMR code will not destroy the current experiment but needs it open to extract the user path information.
There are three ways to launch your workflow:
- Keeping the editor open:
Hit [Execute] button at the top of the window. It is safe to close the Editor window at this point - it will not affect KovriginNMR operation.
- Close the Editor and type the name of your workflow (without '.py') on a command line of Topspin. Hit [Enter] on a keyboard and your script starts.
- Open Workflow: Open Python Browser, select your file and click Execute button on the bottom of the window.
NOTE: Typos in your script will stop its execution (trigger an error). An effective way of quickly checking the code is running it with NMR_Workflow session created with an option run_mode=MAKE_EXPTS_ONLY. This will have entire script execute except for commands controlling the spectrometer and, thus, verify that there are no typos or errors in the code.
When NMR_Workflow object is created in a KovriginNMR script, it creates an HTML report that may be viewed in any browser and places it next to the data on your drive. To view the last created report, click Show Most Recent Report button on KovriginNMR toolbar:
The report will contain all info that you entered when creating NMR experiments in KovriginNMR and all commands along with time stamps during the script operation. For example, below I added six samples to the automatic sample changer database, created a PROTON experiment and then also added an arbitrary text to be displayed along.
The HTML report file is updated on a disk as the workflow proceeds but the browser window does not update automatically. Therefore, to click Refresh button (or its equivalent) periodically to reload the browser window from a disk.
Aborting KovriginNMR operation
If you realized something wrong is going on you may decide to abort operation of KovriginNMR. To this, click [Workflow Control] button and then select [Stop Workflow - create STOP signal]. The action will not be immediate: when KovriginNMR launches different Topspin commands like atma, zg, topshim, etc., it has to wait for their completion. The [Stop Workflow - create STOP signal] button creates a "stop signal" for the KovriginNMR code, and it will exit as soon as the programming control is returned from Topspin back to Python. You will be reminded about this with a message:
Click Close to proceed.
When the current process finishes and returns control to KovriginNMR, it aborts your script and shows you a confirmation window:
If you have a long acquisition currently running, the KovriginNMR script will have to wait for zg command to finish. If you don't need this data you may click Stop button on Topspin standard toolbar or issue stop on a Topspin command line. Shimming may be interrupted by issuing topshim stop. There is no a reliable way to stop atma - I suggest to wait till it finishes itself to avoid hardware errors.
Another sign that Python operation stopped is a message in the status bar (for my ek_test1.py in this example):
Pausing: Temporarily suspending KovriginNMR operation
If you need to suspend operation of KovriginNMR with intention to resume it, you should click [Workflow Control] and select [Pause Workflow: create a PAUSE signal]. Same as with the stop signal, the effect may be not immediate. The KovriginNMR will pause as soon as Topspin returns control to Python and confirm that with this message:
Please, read its content - it is very important. You may do whatever you want during this "KovriginNMR pause" but you must bring everything back as it was before you want to resume. Just imagine you working on a spectrometer and somebody calls you up away from the room. You pause your operation and go. During that time another person comes to the workstation and uses the instrument. Unless they bring EVERYTHING back to where it was - your subsequent operation will be compromised. KovriginNMR will attempt to restore Topspin window to the same open experiment but you MUST make sure the same sample is inserted, and the probe is tuned and shimmed!!!
To resume KovriginNMR script, click [Workflow Control] and select [Resume Workflow: Remove PAUSE and STOP signals]. A message box confirms that KovriginNMR code resumed:
The KovriginNMR code does not wait for you to hit [Close], it already runs.
Aborting while paused
While in the pause, you may hit [Stop Workflow - create STOP signal] and abort KovriginNMR script altogether.
IMPORTANT: KovriginNMR runs inside Topspin. When new data sets are created or any operations are performed on data sets, they will be actually displayed in the Topspin window. If you accidentally or intentionally display a different experiment in the Topspin window in the middle of the KovriginNMR operation, the results will be unpredictable. Therefore, the recommended practice is to dedicate the Topspin window to operation of KovriginNMR and do not do anything in it until the workflow is finished.
What if I want to look at the data or temperature or else?
To do something else in Topspin while KovriginNMR script is running, you must open a new Topspin window from KovriginNMR toolbar by clicking [New Topspin] button:
The original Topspin window must remain exclusively under KovriginNMR control while the new one may be utilized for any other purposes: for example, inspecting the acquired experimental data sets or monitoring temperature of the spectrometer. In the example below, I arranged the original window on the left and the new window on the right. You can recognize the second window by a symbol (2) in the window title. I opened EDTE in the second window and may open some dataset for processing (all while KovriginNMR runs).
NOTE: Do not open the dataset that is being currently acquired - you might interfere with KovriginNMR. The dataset that has already finished acquisition is safe to open. Please, consult the HTML report to know what datasets have already finished acquisition.
This command is equivalent to 'newtop' issued on a command line of Topspin.
Debugging the code, and monitoring execution progress
To check values of variables and make checkpoints at different parts of your code, follow these instructions: Debugging and checking values of variables
An brief introduction to Python in Topspin is found in Topspin manual Python Tutorial in section. Help:Manuals (docs): Programming Manuals section. Below I show general steps of handling Python errors.
General structure of error report
When I am trying to start my script (ek_test.py in this example) while it contains a Python synthax or other programming error, the Topspin brings up a generic error message box:
To find out what the error is, I click on [Details] button and a text window opens. This window contains three sections as shown below:
The error description is in the middle section (by unknown reason, it is shown twice). In the case above, the error is formally triggered by line 20 of my script ek_test.py, after 2 positions from the left:
SyntaxError: ("mismatched input '' expecting EOF", ('/opt/topspin4.1.3/exp/stan/nmr/py/user/ek_test.py', 20, 2, ' MSG("Please Click CLOSE to continue", "MSG Test")\n'))Often the error description tells you what happened but sometimes it is quite cryptic. In the example above, it complains about two extra spaces that I added in front of one of lines of my Python code block. Please, see examples below on how to decifer the Python error messages.
NOTE: When you are reaching out to someone for help, you should copy-paste this error message into email for them to read, in addition to your explanations of what is not working.
Examples of errors
Mismatched input: uneven indentation in the code block
Here, I will write a short script and trigger some errors for you to illustrate what to look for. Let's assume my code is this:
def test_function(x):
return x
a=1
b=test_function(a)
c=a+b
I intentionally set the code at the edge of the screen for you to clearly see indentation pattern of the Python code lines: they all start with the same (first) position of each line (except for the inside of the function definition). Now, I will break the Python syntax rule that all lines in a block must be identically indented, and displace one of the lines to the right by inserting one space character:
def test_function(x):
return x
a=1
b=test_function(a)
c=a+b
This triggers an error message:
"SyntaxError: ("mismatched input '' expecting EOF", ('/opt/topspin4.1.3/exp/stan/nmr/py/user/py-test.py', 5, 1, ' b=test_function(a)\n'))"The message says that the Python interpreter encountered a SyntaxError in py-test.py, which is related to a mismatched input on a line 5, one character from the beginning of the line (content of which is also shown). That means, Python detected my extra space!
Such extra spaces breaking identical intentation in the code block are often a problem, because one space is enough to trigger an error, but it is not very visible in the script text. This situation often happens when you comment or uncomment lines of code.
NOTE: This issue may also occur when you are preparing your code in a different editor (like MS Word) or copying from a web browser. Some editors automatically add spaces or tab characters where they deem appropriate for a pretty look of the document. This, however, will trigger a Python error because it will create a uneven indentation. If you want to use an external editor for creating your code, make sure this editor is intended for computer programming, which means it does not do autocorrection and does not insert spaces, tabs, and special characters by itself.
NOTE 2: Sometimes external editors will also use special formatting symbols inside your text, which are not visible in the text. However, they will be visible for the Python interpreter and will trigger an error. You should take care not to use such editors (see previous note). If you got into such situation when your code brought from a different editor triggers a syntax error yet you have no visible error, you, may try
- commenting out the line that triggers the problem and see if the error disappears;
- deleting the line entirely (thus getting rid of invisible characters) and retyping it manually on a keyboard.
Typo in a variable name
The next error that I will trigger is a typo in a variable name: instead of a I will type in aa in the line 5:
def test_function(x): return x a=1 b=test_function(aa) c=b+a
In my code, line 5 calls for a variable aa, which has not been ever assigned because it has not been meant to exist in the first place. The Traceback message error is a little more comprehensible now:
Traceback (most recent call last): File "/opt/topspin4.1.3/exp/stan/nmr/py/user/py-test.py", line 5, in <module> b=test_function(aa) NameError: name 'aa' is not definedNameError is an indication that you are using a name for a variable or function, which is not known to Python from your script. It happens due to typos or if the variable has not been assigned (received a value) in the previous lines of code.
Typo inside a function
Next level of complexity interpreting error messages comes when an error happens inside the function, which is called from somewhere else in the code. For example, I will missspell a variable name inside my test_function, and type xx instead of x on line 2:
def test_function(x):
return xx
a=1
b=test_function(a)
c=b+a
The traceback now contains two lines involved, line 2 and line 5:
Traceback (most recent call last):
File "/opt/topspin4.1.3/exp/stan/nmr/py/user/py-test.py", line 5, in <module>
b=test_function(a)
File "/opt/topspin4.1.3/exp/stan/nmr/py/user/py-test.py", line 2, in test_function
return xx
NameError: global name 'xx' is not definedThis sequence of lines is called an error stack. You start looking at it from the bottom: line 2 in my file is where the error occurred originally. The line 5 error is recorded just because is it this line that calls the function where error occurred.
Computation error, run-time
Computation error or run-time error occurs when some value in your calculations is incorrect leading to a failure in the code execution. The error message will not be triggered until the calculation is actually done. Tracing the run-tume error back to the source is more difficult. Generally, such errors are failures of your algorithm. Below, I will trigger a runtime error by division by zero. Let's try this code:
def test_function(x): return 1/x a=0 b=test_function(a) c=b+a
Here I will assign zero to a on line 4 and pass it to my test_function on line 5. The division by zero happens inside the function code block, that is at the line 2. Here is the error stack for this situation:
Traceback (most recent call last): File "/opt/topspin4.1.3/exp/stan/nmr/py/user/py-test.py", line 5, in <module> b=test_function(a) File "/opt/topspin4.1.3/exp/stan/nmr/py/user/py-test.py", line 2, in test_function return 1/x ZeroDivisionError: integer division or modulo by zeroThe last message of the error stack indicates the line 2 as the place where a division by zero occurs. If you walk up the stack to a previous message, which is about line 5, you see that it was a call of test_function with a variable a. We know that the source of this error is actually in the line 4, where zero was assigned to a variable a.
Runtime computation errors are most difficult to detect. The common approach is to change values of variables in a systematic way and observe a response of your script.
To be able to print the run-time values of variables in Python in Topspin, follow these guidelines: Practical Examples:Debugging and checking values of variables
Evgenii Kovrigin (C) 2022