U
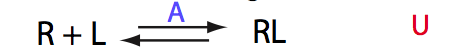
U
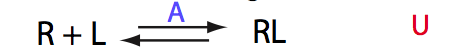
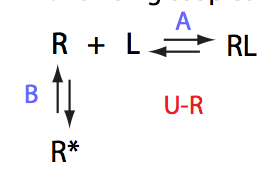
U_R
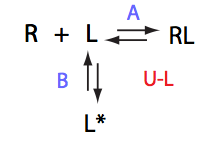
U_L
U_RL
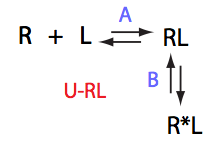
U_RL

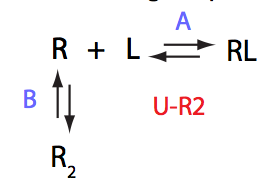
U_R2
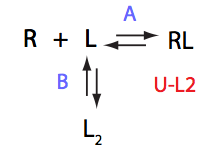
U_L2
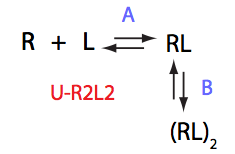
U_R2L2
U_R2_L2

Goal of this section is to present cases how line shapes for different models will appear in direct titrations: L(titrant) to R(NMR-observable); as well as in reverse titrations: R(unlabeled titrant) to L(NMR-observable). I want to explore opportunities to tell apart different mechanisms, which otherwise produce very similar line shapes.
We will consider models that may be alternatives of each other:
| Direct titration: Ligand added to NMR-observable Receptor | Reverse titration : Receptor added to NMR-observable Ligand | ||
| 1 | U
|
U
|
|
| 2 | U_R
|
U_L
|
|
| 3 | U_RL
|
U_RL
|
|
| 4 | U_R2
|
U_L2
|
|
| 5 | U_R2L2
|
U_R2_L2
|
|
I will consider different combinations of exchange regimes. They are not entirely distinct and better be thought of representation of a continuum of conditions. I want to observe general trends and formulate some recipes on how to analyze these systems.
Additional note: the choice of chemical shifts for species is arbitrary. I only try to follow general relations between Kex and dw but it is not always unambiguous.
unfinished
Back to LineShapeKin Simulation Tutorial