Fast isomerization removes any opportunity to detect multiple species. We will rely on fitting to the 2-site modela as a diagnostic tool.
Common parameters we use are:
# Association constants
Ka_names A B
Ka 1e8 5
# Rate constants of REVERSE reactions
k_names A B
k2 1 200

| between peaks/dw(B) | 50/25 | 100/50 | 200/100 | 300/150 | 300/50 | 50/300 |
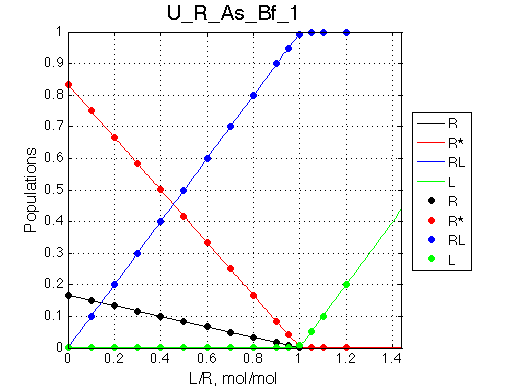
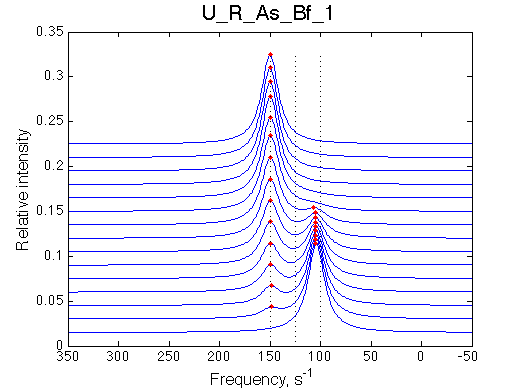
Simulate setup U_R_As_Bf_1 |
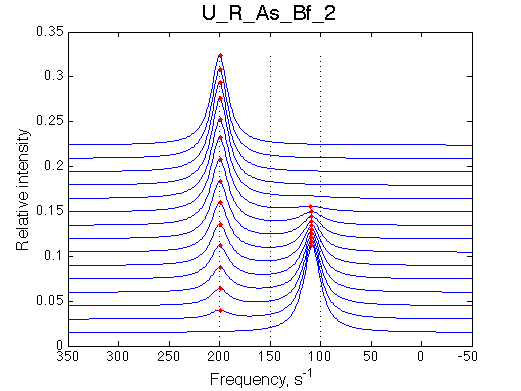
Simulate setup U_R_As_Bf_2 |
Simulate setup U_R_As_Bf_3 |
Simulate setup U_R_As_Bf_4 |
Simulate setup U_R_As_Bf_5 |
Simulate setup U_R_As_Bf_6 | |
 |
 |
 |
 |
 |
 |
|
 |
 |
 |
 |
 |
 |
|
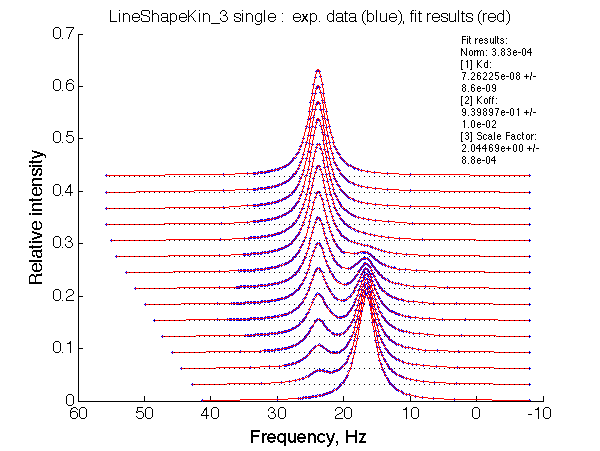
| Optimum norm: 3.83e-04 [1] Kd: 7.26225e-08 +/- 8.6e-09 [2] Koff: 9.39897e-01 +/- 1.0e-02 [3] Scale Factor: 2.04469e+00 +/- 8.8e-04 |
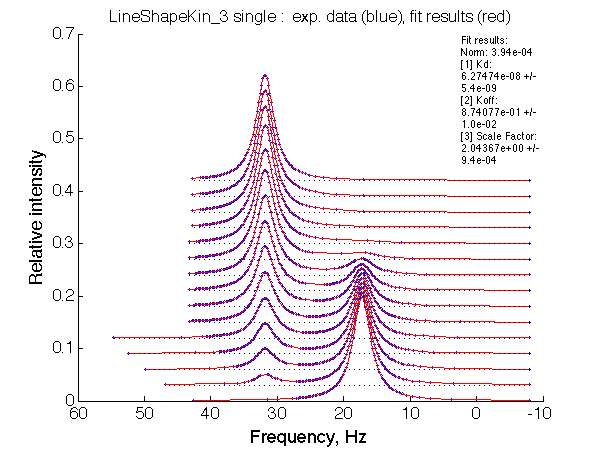
Optimum norm: 3.94e-04 [1] Kd: 6.27474e-08 +/- 5.4e-09 [2] Koff: 8.74077e-01 +/- 1.0e-02 [3] Scale Factor: 2.04367e+00 +/- 9.4e-04 |
Optimum norm: 1.16e-03 [1] Kd: 4.67171e-08 +/- 6.9e-09 [2] Koff: 8.05507e-01 +/- 1.9e-02 [3] Scale Factor: 2.02144e+00 +/- 1.8e-03 |
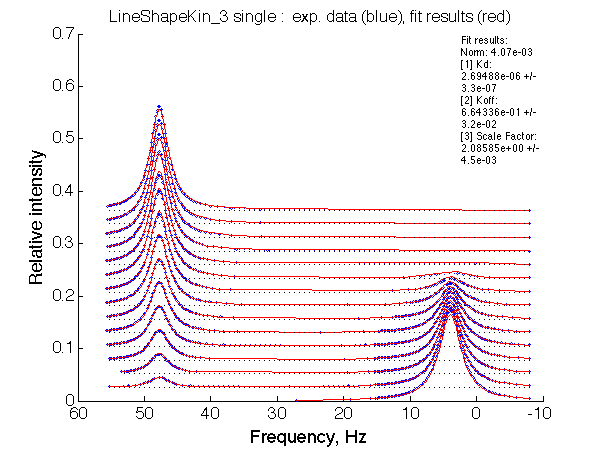
Optimum norm: 4.07e-03 [1] Kd: 2.69488e-06 +/- 3.3e-07 [2] Koff: 6.64336e-01 +/- 3.2e-02 [3] Scale Factor: 2.08585e+00 +/- 4.5e-03 |
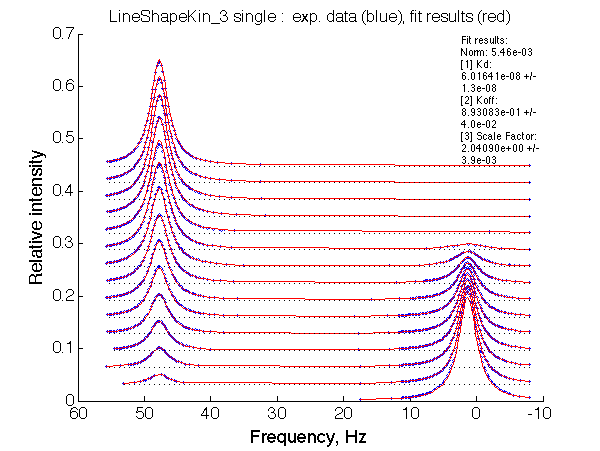
Optimum norm: 5.46e-03 [1] Kd: 6.01641e-08 +/- 1.3e-08 [2] Koff: 8.93083e-01 +/- 4.0e-02 [3] Scale Factor: 2.04090e+00 +/- 3.9e-03 |
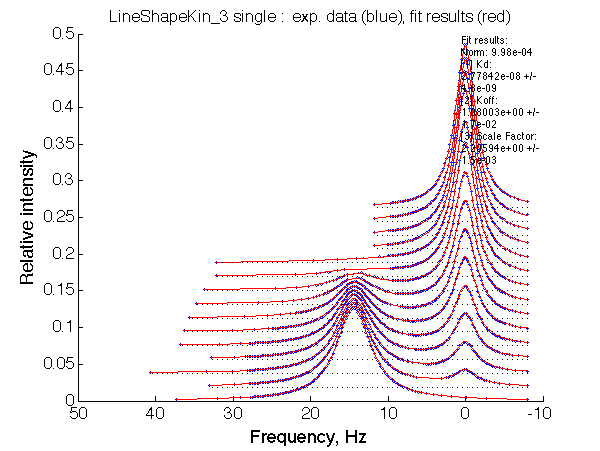
ptimum norm: 9.98e-04 [1] Kd: 2.77842e-08 +/- 4.8e-09 [2] Koff: 1.08003e+00 +/- 1.7e-02 [3] Scale Factor: 2.20594e+00 +/- 1.5e-03 |
No apparent dependence of fitted Kd on the offset.
Koff drops with combined dw of isomerization and binding.
This is a weak effect.
Back to Analysis of Direct and Reverse Titrations