ProjectName:="B_R2_R2L2";
CurrentPath:="/Users/kovrigin/Documents/Workspace/Data/Data.XV/EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/B_R2_R2L2_model/";
![]()
![]()
B_R2_R2L2
Receptor dimerization coupled with binding of one ligand per receptor monomer
2. Basic equilibrium equations
3. Analysis of statistical effects on binding constants
4. Derivation of equations for concentrations of species in terms of MACROSCOPIC constants
Summary of equations for equilibrium concentrations and relationship to MICROSCOPIC constants
5. Prepare equations for a numeric solution in MICROSCOPIC constants
6. Save results on disk for future use
In this notebook I will derive equations for binding of a ligand to a receptor molecule coupled with dimerization of the receptor and prepare equations for numerical simulations.
Analysis will be done in
EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/B-R2-R2L2_model/B_R2_R2L2_analysis.mn
clean up workspace
reset()
Set path to save results into:
ProjectName:="B_R2_R2L2";
CurrentPath:="/Users/kovrigin/Documents/Workspace/Data/Data.XV/EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/B_R2_R2L2_model/";
![]()
![]()
We need to consider the system two ways: using macroscopic and microscopic binding constants [1]. Overall, these are two completely equivalent treatments but interpretation in terms of mechanistic events is only available for microscopic constants. We are especially interested in microscopic constants because we may assess cooperativity between the sites.
Symmetry of the system makes it easy to use macroscopic treatment and simply substitute microscopic constants into it.

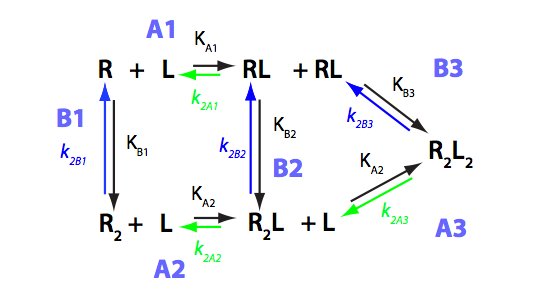
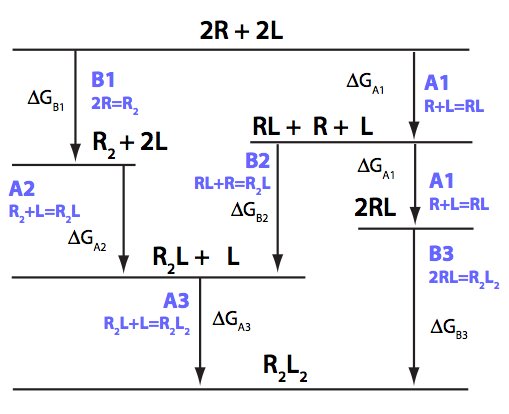
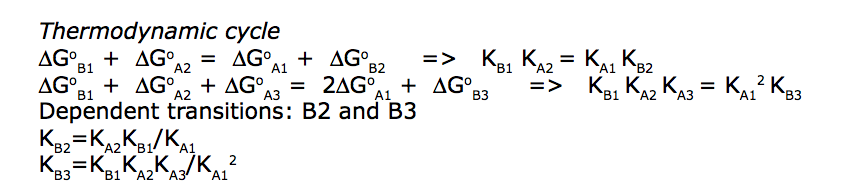

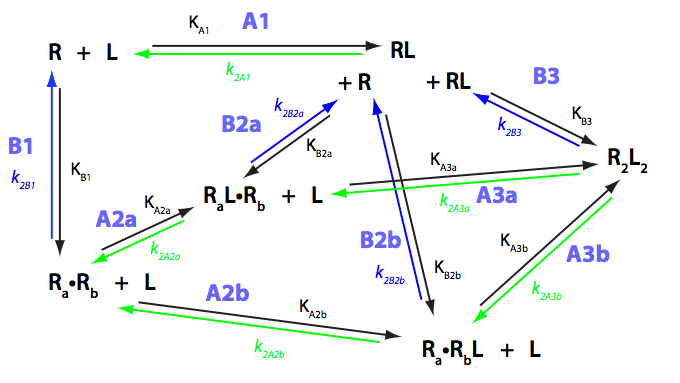
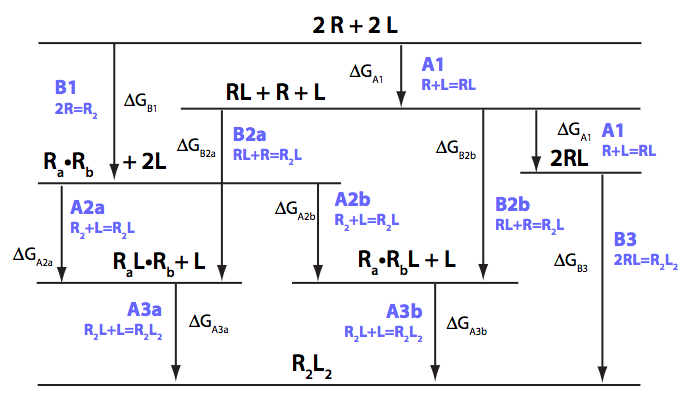
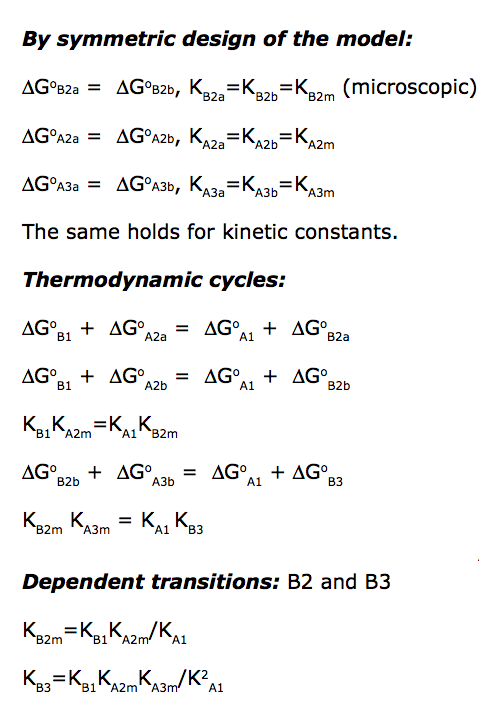
Binding and isomerization constants
All binding constants I am using are association constants.
These relationships serve as restraints for solve(), but not restrict these values in calculations!
Macroscopic constants
K_A_1
K_A_1 ;
assumeAlso(K_A_1 > 0):
assumeAlso(K_A_1 , R_):
![]()
K_A_2
K_A_2 ;
assumeAlso(K_A_2 > 0):
assumeAlso(K_A_2 , R_):
![]()
K_A_3
K_A_3 ;
assumeAlso(K_A_3 > 0):
assumeAlso(K_A_3 , R_):
![]()
K_B_1
K_B_1 ;
assumeAlso(K_B_1 > 0):
assumeAlso(K_B_1 , R_):
![]()
K_B_2
K_B_2 ;
assumeAlso(K_B_2 > 0):
assumeAlso(K_B_2 , R_):
![]()
K_B_3
K_B_3 ;
assumeAlso(K_B_3 > 0):
assumeAlso(K_B_3 , R_):
![]()
Microscopic constants
KB2micro
K_B_2_m;
assumeAlso(K_B_2_m > 0):
assumeAlso(K_B_2_m , R_):
![]()
KA2micro
K_A_2_m;
assumeAlso(K_A_2_m > 0):
assumeAlso(K_A_2_m , R_):
![]()
KA3micro
K_A_3_m;
assumeAlso(K_A_3_m > 0):
assumeAlso(K_A_3_m , R_):
![]()
Total concentrations
Rtot - total concentration of the receptor
Rtot;
assumeAlso(Rtot>0):
assumeAlso(Rtot,R_):
![]()
Ltot - total concentration of a ligand
Ltot;
assumeAlso(Ltot>0):
assumeAlso(Ltot,R_):
![]()
Equilibrium concentrations
Req
Req;
assumeAlso(Req>0):
assumeAlso(Req<Rtot):
assumeAlso(Req,R_):
![]()
Leq
Leq;
assumeAlso(Leq>0):
assumeAlso(Leq<Ltot):
assumeAlso(Leq,R_):
![]()
RLeq
RLeq;
assumeAlso(RLeq>0):
assumeAlso(RLeq<Rtot):
assumeAlso(RLeq<Ltot):
assumeAlso(RLeq,R_):
![]()
R2eq
R2eq;
assumeAlso(R2eq>0):
assumeAlso(R2eq<Rtot/2):
assumeAlso(R2eq,R_):
![]()
R2Leq
R2Leq;
assumeAlso(R2Leq>0):
assumeAlso(R2Leq<Rtot/2):
assumeAlso(R2Leq<Ltot):
assumeAlso(R2Leq,R_):
![]()
R2L2eq
R2L2eq;
assumeAlso(R2L2eq>0):
assumeAlso(R2L2eq<Rtot/2):
assumeAlso(R2L2eq<Ltot/2):
assumeAlso(R2L2eq,R_):
![]()
Microscopic species
R2Laeq;
assumeAlso(R2Laeq>0):
assumeAlso(R2Laeq<Rtot/2):
assumeAlso(R2Laeq<Ltot):
assumeAlso(R2Laeq,R_):
![]()
R2Lbeq;
assumeAlso(R2Lbeq>0):
assumeAlso(R2Lbeq<Rtot/2):
assumeAlso(R2Lbeq<Ltot):
assumeAlso(R2Lbeq,R_):
![]()
anames(Properties,User);

2. Basic equilibrium equations
Working equation: I will try to express analytical [L] from equation for a total concentration of a receptor or use the expression for a numeric solution if analytical is not possible.
Total concentrations of a receptor and a ligand
eq2_1:= Rtot = Req + RLeq + 2*R2eq + 2*R2Leq + 2*R2L2eq;
![]()
eq2_2:= Ltot = Leq + RLeq + R2Leq + 2*R2L2eq;
![]()
Write equilibrium thermodynamics equations using macroscopic constants (KB2 and KB3 are chosen to be dependent constants).
eq2_3:= K_A_1 = RLeq / (Req*Leq);
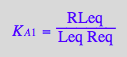
eq2_4:= K_A_2 = R2Leq / (R2eq*Leq);
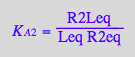
eq2_5:= K_A_3 = R2L2eq / (R2Leq*Leq);
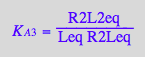
eq2_6:= K_B_1 = R2eq / (Req*Req);
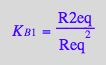
eq2_7:= K_B_2 = R2Leq / (RLeq*Req);
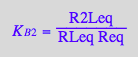
eq2_8:= K_B_3 = R2L2eq / (RLeq*RLeq);
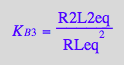
Write additional equilibrium equations defining microscopic constants and species
eq2_9:= R2Leq = R2Laeq + R2Lbeq
![]()
eq2_10:= K_A_2_a = R2Laeq / (R2eq*Leq)
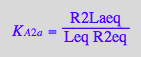
eq2_11:= K_A_2_b = R2Lbeq / (R2eq*Leq)
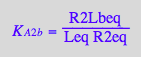
eq2_12:= K_A_3_a = R2L2eq / (R2Laeq*Leq)
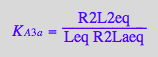
eq2_13:= K_A_3_b = R2L2eq / (R2Lbeq*Leq)
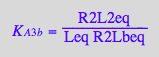
eq2_14:= K_B_2_a = R2Laeq / (RLeq*Req)
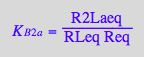
eq2_15:= K_B_2_b = R2Lbeq / (RLeq*Req)
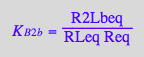
Equalities for microscopic constants due to symmetry of the model:
eq2_16:= K_B_2_a = K_B_2_m;
eq2_17:= K_B_2_b = K_B_2_m;
eq2_18:= K_A_2_a = K_A_2_m;
eq2_19:= K_A_2_b = K_A_2_m;
eq2_20:= K_A_3_a = K_A_3_m;
eq2_21:= K_A_3_b = K_A_3_m;
![]()
![]()
![]()
![]()
![]()
![]()
3. Analysis of statistical effects on binding constants
Establish relationship between microscopic and macroscopic binding constants
B2 transition
R2Laeq
eq2_14;
solve(%,R2Laeq);
%[1][1];
eq3_1:= R2Laeq = %
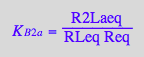

![]()
![]()
R2Lbeq
eq2_15;
solve(%,R2Lbeq);
%[1][1];
eq3_2:= R2Lbeq = %
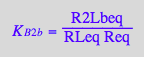

![]()
![]()
Substitute to mass conservation law
eq2_9;
% | eq3_1 | eq3_2;
eq3_3:= %:
![]()
![]()
Substitute in K_B_2 expression
eq2_7;
% | eq3_3;
normal(%);
% | eq2_16 | eq2_17;
eq3_4a:= %:
solve(%,K_B_2_m);
eq3_4b:= K_B_2_m = %[1]
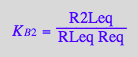
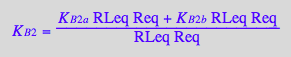
![]()
![]()

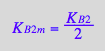
A2 transition
R2Laeq
eq2_10;
solve(%,R2Laeq);
%[1][1];
eq3_5:= R2Laeq = %;
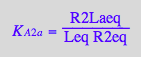

![]()
![]()
R2Lbeq
eq2_11;
solve(%,R2Lbeq);
%[1][1];
eq3_6:= R2Lbeq = %
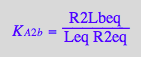

![]()
![]()
Mass conservation
eq2_9;
% | eq3_5 | eq3_6;
eq3_7:= %:
![]()
![]()
Substitute into macroscopic constant equation
eq2_4;
% | eq3_7 ;
% | eq2_18 | eq2_19;
eq3_9a:= %:
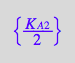
solve(%,K_A_2_m);
eq3_9b:= K_A_2_m = %[1]
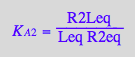
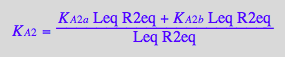
![]()
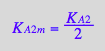
A3 transition
R2Laeq
eq2_12;
solve(%,R2Laeq);
%[1][1];
eq3_10:= R2Laeq = %
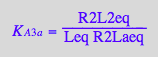


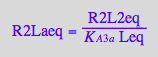
R2Lbeq
eq2_13;
solve(%, R2Lbeq);
%[1][1];
eq3_11:= R2Lbeq = %
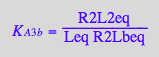


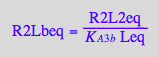
Mass conservation
eq2_9;
% | eq3_10 | eq3_11;
eq3_12:= %:
![]()
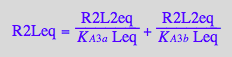
Substitute into the macroscopic constants equation
eq2_5;
% | eq3_12;
% | eq2_20 | eq2_21;
eq3_13a:= %:
solve(%,K_A_3_m);
eq3_13b:= K_A_3_m = %[1]
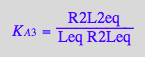
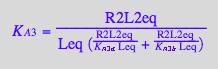
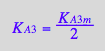
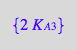
![]()
Relationships between kinetic and equilibrium constants
A2 transition
Macro:
dR2/dt = -k_1_A_2*R2eq*Leq + k_2_A_2*R2Leq
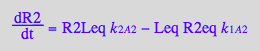
Micro:
dRaRb/dt = -2*k_1_A_2_m*R2eq*Leq + k_2_A_2_m*RaLRbeq +k_2_A_2_m*RaRbLeq

dRaRb/dt= -2*k_1_A_2_m*R2eq*Leq + k_2_A_2_m*R2Leq
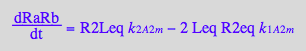
Comparing first and last equation we can equate:
eq3_20:= k_2_A_2_m=k_2_A_2
![]()
eq3_21:= 2*k_1_A_2_m=k_1_A_2
![]()
A3 transition
Macro:
dR2L2/dt = k_1_A_3*R2Leq*Leq - k_2_A_3*R2L2eq
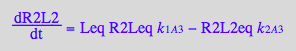
Micro:
dR2L2/dt = -2*k_2_A_3_m*R2L2eq + k_1_A_3_m*RaLRbeq*Leq +k_1_A_3_m*RaRbLeq*Leq

dR2L2/dt= -2*k_2_A_3_m*R2L2eq + k_1_A_3_m*R2Leq*Leq
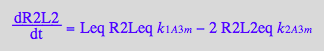
Comparing first and last equations we can equate:
eq3_22:= k_1_A_3_m=k_1_A_3
![]()
eq3_23:= 2*k_2_A_3_m=k_2_A_3
![]()
Summary of relationships between
micro- and macroscopic equilibrium binding constants
eq3_4a; eq3_4b;
![]()
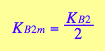
eq3_9a; eq3_9b
![]()
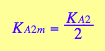
eq3_13a; eq3_13b
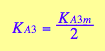
![]()
Relationships of kinetic constants
eq3_20;
![]()
eq3_21
![]()
eq3_22
![]()
eq3_23
![]()
4. Derivation of equations for concentrations of species in terms of MACROSCOPIC constants
Express Leq as a function of macroscopic constants and total concentrations of a receptor and a ligand
Independent equations to use:
eq2_1;eq2_2
![]()
![]()
eq2_3;eq2_4;eq2_5;eq2_6;eq3_9a;eq3_13a
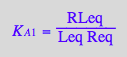
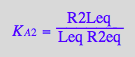
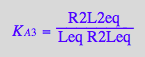
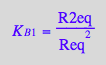
![]()
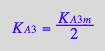
Express complex species concentrations:
RLeq
eq2_3;
solve(%,RLeq);
%[1][1];
eq4_1:= RLeq = %
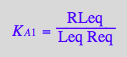
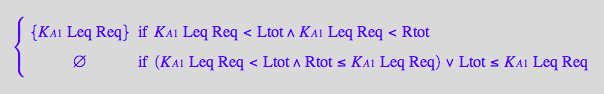
![]()
![]()
R2eq
eq2_6;
solve(%,R2eq);
%[1][1];
eq4_2:= R2eq = %
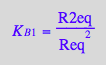
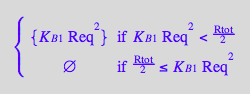
![]()
![]()
R2Leq
eq2_4;
% | eq4_2;
solve(%,R2Leq);
%[1][1];
eq4_3:= R2Leq = % | eq3_9a
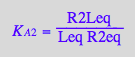

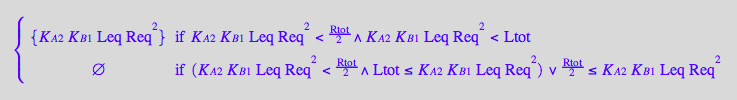
![]()
![]()
R2L2eq
eq2_5;
% | eq4_3;
solve(%, R2L2eq);
%[1][1];
eq4_4:= R2L2eq = % | eq3_13a
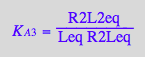

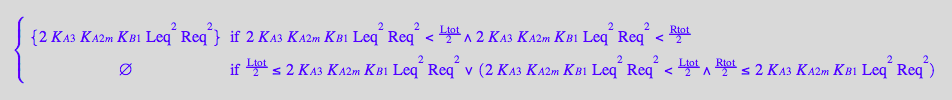
![]()
![]()
Substitute into a mass conservation law for L and express Req
eq2_2;
% | eq4_4;
% | eq4_3;
% | eq4_1;
eq4_5:= %:
![]()
![]()
![]()
![]()
Solve for Req
solutions4_5:=solve(eq4_5,Req):
eq4_6:= solutions4_5[i,1] $ i=1..nops(solutions4_5):
print(Unquoted,"Number of formal solutions: ".nops(%))
Number of formal solutions: 4
How many independent soltions we have?
Is 1st solution a combination of 2nd and 3rd?
solution1:=eq4_6[1]; // a sequence of roots (multiple solutions)
solution2:=eq4_6[2][1]; // extract equation out of a sequence
solution3:=eq4_6[3][1]; // extract equation out of a sequence
if solution2 in solution1
then print(Unquoted,"First set of roots contains the second root.");
else print(Unquoted,"First set of roots DOES NOT contain the second root!");
end_if;
if solution3 in solution1
then print(Unquoted,"First set of roots contains the third root.");
else print(Unquoted,"First set of roots DOES NOT contain the third root!");
end_if;
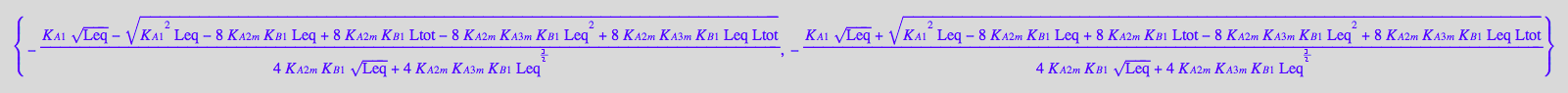
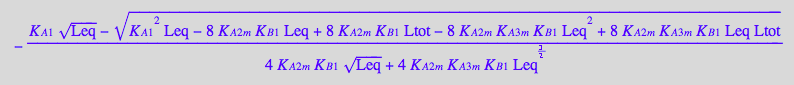
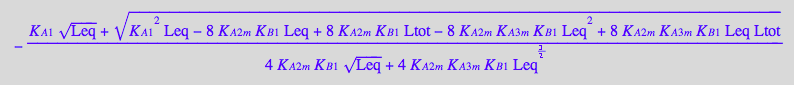
First set of roots contains the second root.
First set of roots contains the third root.
Check correctness of the solutions by substitution into original equation solved:
Check first root
test1:= eq4_5 | Req=solution2:
normal(%);
![]()
Check the second root
test2:= eq4_5 | Req=solution3:
normal(%);
![]()
Both solutions are correct.
Choose meaningful one for further work
solution2 | Rtot=1 | Ltot=0.1 | Leq=0.001 | K_A_1=1 | K_B_1=1 | K_A_2_m=1 | K_A_3_m=1
![]()
solution3 | Rtot=0.1 | Ltot=0.1 | Leq=0.01 | K_A_1=1 | K_B_1=1 | K_A_2_m=1 | K_A_3_m=1
![]()
Solution 2 is meaningful !
ASSIGN TO AN EQUATION:
eq4_7:= Req = solution2;
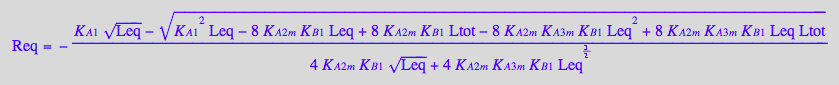
Finally, substitute all into a mass conservation law for a receptor
eq2_1;
% | eq4_1 | eq4_2 | eq4_3 | eq4_4 ;
% | eq4_7;
eq4_8:= %:
![]()
![]()

Check consistency: derive equations for dependent constants
Kb2
eq2_7 ;
% | eq4_3 | eq4_1 ;
solve(%,K_B_2);
eq4_9:= K_B_2 =%[1]
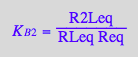
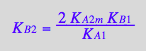
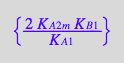
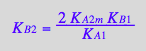
Correct.
KB3
eq2_8;
% | eq4_1;
% | eq4_4;
eq4_10:= %:
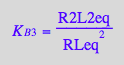
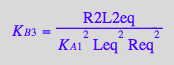

Summary of equations for equilibrium concentrations
Independent parameters:
Rtot, Ltot, K_A_1, K_B_1, K_A_2, K_A_3
![]()
Macroscopic constants in terms of microscopic ones:
eq3_9a
![]()
eq3_4a
![]()
eq3_13a
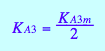
Relationships of kinetic constants
eq3_20;
![]()
eq3_21
![]()
eq3_22
![]()
eq3_23
![]()
Equation to solve:
Rtot=f([L])
eq4_8

Equilibrium concentrations of species:
[R]
eq4_7
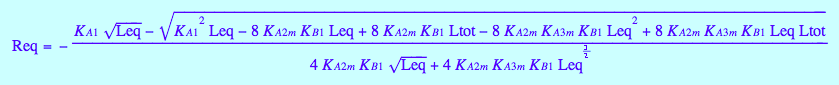
[RL]
eq4_1
![]()
[R2]
eq4_2
![]()
[R2L]
eq4_3
![]()
[R2L2]
eq4_4
![]()
Dependent equilibrium constants
eq4_9
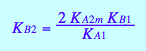
eq4_10

5. Prepare equations for a numeric solution in MICROSCOPIC constants
I will express all equilibrium concentrations in terms of each other in successive order, easy to code in Matlab.
Make a function for numeric solving and switch to LRratio=Ltot/Rtot
eq5_1:= Ltot=LRratio*Rtot;
fRtot_B_R2_R2L2:=(Rtot, LRratio, Leq, K_A_1, K_B_1, K_A_2_m, K_A_3_m) --> eq4_8[2] | eq5_1;
![]()

Assume some constant values for testing
Total_R:=1e-3:
Ka1:=1e7:
Kb1:=2e3:
Ka2m:=1e5:
Ka3m:=1e5:
LR_Ratio_max:=2.5:
LR_Ratio:=0.8:
Test fRtot:
fRtot_B_R2_R2L2(Total_R, LR_Ratio, 1e-4, Ka1, Kb1, Ka2m, Ka3m)
![]()
it works OK.
Define a procedure for numeric solving the equation Rtot=f([L]) for [L] thus creating a function [L]=f(Rtot,...)
// WARNING: make sure the Leq search range starts with a non-zero number!!!! Use a number larger than that to create approximation of LRratio=0
pnLeq_B_R2_R2L2:= proc(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m)
/* Parameter names should be different from
variable names used in the equation!!!
If you see an error message:
"Error: Illegal operand [_index];
during evaluation of 'your function name'"
it means fsolve() returned FAIL and you need to check
values of all parameters passed to the function fRtot
*/
local result, L;
begin
//print(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
// numeric solving equation for Leq in a restricted range.
// WARNING: make sure the range starts with a non-zero number!!!!
result:=numeric::fsolve(
Total_R=fRtot_B_R2_R2L2(Total_R, LR_Ratio, Leq, Ka1, Kb1, Ka2m, Ka3m),
Leq=10e-32..Total_R*LR_Ratio);
//print(result);
// extract answer from equation
L:=result[1][2];
//print(Unquoted,"solve() residual: ".(Total_R-fRtot_B_R2_R2L2(Total_R, LR_Ratio, L, Ka1, Kb1, Ka2m, Ka3m)));
return(L);
end_proc;
![]()
test operation
Total_R:=1:
Ka1:=1e7:
Kb1:=2e3:
Ka2m:=1e5:
Ka3m:=1e5:
LR_Ratio_max:=2.5:
LR_Ratio:=10:
DIGITS:=10;
pnLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) ;
![]()
![]()
Make "portable" equations
(unique names for future use):
eq3_9a
![]()
eq3_4a
![]()
eq3_13a
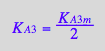
RLeq_B_R2_R2L2:=eq4_1;
R2eq_B_R2_R2L2:=eq4_2;
R2Leq_B_R2_R2L2:=eq4_3;
R2L2eq_B_R2_R2L2:=eq4_4;
Req_B_R2_R2L2:=eq4_7;
Rtot_B_R2_R2L2:=eq4_8:
KB2_B_R2_R2L2:=eq4_9;
KB3_B_R2_R2L2:=eq4_10;
KA2_B_R2_R2L2:=eq3_9a;
KB2_B_R2_R2L2:=eq3_4a;
KA3_B_R2_R2L2:=eq3_13a;
![]()
![]()
![]()
![]()
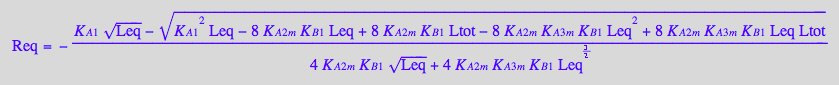
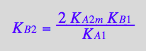

![]()
![]()
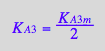
Define functions for equilibrium concentrations of all species:
[R]
Req_B_R2_R2L2;
pnReq_B_R2_R2L2:= proc(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m)
// Parameter names should be different from
// variable names used in the equation!!!
local L;
begin
L:=pnLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
// Equation for [R]
Req_B_R2_R2L2[2] | Ltot=Total_R*LR_Ratio | Leq=L | K_A_1=Ka1 | K_B_1=Kb1 | K_A_2_m=Ka2m | K_A_3_m=Ka3m;
end_proc
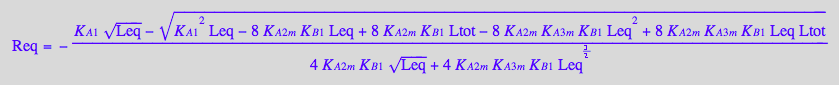
![]()
test
Total_R:=1:
Ka1:=10:
Kb1:=1e3:
Ka2m:=1:
Ka3m:=1:
LR_Ratio_min:=1e-2:
LR_Ratio_max:=10:
pnReq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m) ;
pnReq_B_R2_R2L2(Total_R, LR_Ratio_max, Ka1, Kb1, Ka2m, Ka3m) ;
![]()
![]()
OK
Total_R:=1:
Ka1:=1.0:
Ka2m:=10.0:
Ka3m:=100.0:
Kb1:=0.1:
LR_Ratio_min:=0.1:
LR_Ratio_max:=1:
pnReq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m) ;
eq4_7[2] | Leq=pnLeq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m) | K_A_1=Ka1 | K_A_2_m=Ka2m | K_A_3_m=Ka3m | K_B_1=Kb1 | Ltot=Total_R*LR_Ratio_min
![]()
![]()
[RL]
RLeq_B_R2_R2L2;
pnRLeq_B_R2_R2L2:= proc(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m)
// Parameter names should be different from
// variable names used in the equation!!!
local L, R;
begin
L:=pnLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
R:=pnReq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
// Equation for [R]
RLeq_B_R2_R2L2[2] | Ltot=Total_R*LR_Ratio | Req=R | Leq=L | K_A_1=Ka1 | K_B_1=Kb1 | K_A_2_m=Ka2m | K_A_3_m=Ka3m;
end_proc
![]()
![]()
test
pnRLeq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m) ;
pnRLeq_B_R2_R2L2(Total_R, LR_Ratio_max, Ka1, Kb1, Ka2m, Ka3m) ;
![]()
![]()
OK
[R2]
R2eq_B_R2_R2L2;
pnR2eq_B_R2_R2L2:= proc(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m)
// Parameter names should be different from
// variable names used in the equation!!!
local L, R;
begin
R:=pnReq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
// Equation for [R]
R2eq_B_R2_R2L2[2] | Ltot=Total_R*LR_Ratio | Req=R | K_A_1=Ka1 | K_B_1=Kb1 | K_A_2_m=Ka2m | K_A_3_m=Ka3m;
end_proc
![]()
![]()
test
pnR2eq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m) ;
pnR2eq_B_R2_R2L2(Total_R, LR_Ratio_max, Ka1, Kb1, Ka2m, Ka3m) ;
![]()
![]()
OK
[R2L]
R2Leq_B_R2_R2L2;
pnR2Leq_B_R2_R2L2:= proc(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m)
// Parameter names should be different from
// variable names used in the equation!!!
local L, R;
begin
L:=pnLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
R:=pnReq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
// Equation for [R]
R2Leq_B_R2_R2L2[2] | Ltot=Total_R*LR_Ratio | Leq=L | Req=R | K_A_1=Ka1 | K_B_1=Kb1 | K_A_2_m=Ka2m | K_A_3_m=Ka3m;
end_proc
![]()
![]()
test
pnR2Leq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m) ;
pnR2Leq_B_R2_R2L2(Total_R, LR_Ratio_max, Ka1, Kb1, Ka2m, Ka3m) ;
![]()
![]()
OK
[R2L2]
R2L2eq_B_R2_R2L2;
pnR2L2eq_B_R2_R2L2:= proc(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m)
// Parameter names should be different from
// variable names used in the equation!!!
local L, R;
begin
L:=pnLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
R:=pnReq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
// Equation for [R]
R2L2eq_B_R2_R2L2[2] | Ltot=Total_R*LR_Ratio | Leq=L | Req=R | K_A_1=Ka1 | K_B_1=Kb1 | K_A_2_m=Ka2m | K_A_3_m=Ka3m;
end_proc
![]()
![]()
test
Total_R:=1:
Ka1:=10:
Kb1:=1e3:
Ka2m:=1:
Ka3m:=1:
LR_Ratio:=8:
pnR2L2eq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
LR_Ratio:=10:
pnR2L2eq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m);
![]()
![]()
OK
Test plotting
Total_R:=1e-3:
Ka1:=10:
Kb1:=1e3:
Ka2m:=1e5:
Ka3m:=1e6:
LR_Ratio_min:=1E-16:
LR_Ratio_max:=1.5:
pnR2L2eq_B_R2_R2L2(Total_R, LR_Ratio_min, Ka1, Kb1, Ka2m, Ka3m);
pnR2L2eq_B_R2_R2L2(Total_R, LR_Ratio_max, Ka1, Kb1, Ka2m, Ka3m);
// Make wrapper functions dependent on LRratio
fnLeq:=LR_Ratio -> pnLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) ;
fnReq:=LR_Ratio -> pnReq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) :
fnRLeq:=LR_Ratio -> pnRLeq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) :
fnR2eq:=LR_Ratio -> pnR2eq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) :
fnR2Leq:=LR_Ratio -> pnR2Leq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) :
fnR2L2eq:=LR_Ratio -> pnR2L2eq_B_R2_R2L2(Total_R, LR_Ratio, Ka1, Kb1, Ka2m, Ka3m) :
print(Unquoted,"Leq ",fnLeq(LR_Ratio_min),fnLeq(LR_Ratio_max));
print(Unquoted,"Req ",fnReq(LR_Ratio_min),fnReq(LR_Ratio_max));
print(Unquoted,"RLeq ",fnLeq(LR_Ratio_min),fnRLeq(LR_Ratio_max));
print(Unquoted,"R2eq ",fnR2eq(LR_Ratio_min),fnR2eq(LR_Ratio_max));
print(Unquoted,"R2Leq ",fnR2Leq(LR_Ratio_min),fnR2Leq(LR_Ratio_max));
print(Unquoted,"R2L2eq ",fnR2L2eq(LR_Ratio_min),fnR2L2eq(LR_Ratio_max));
LW:=1.5:
pLeq:= plot::Function2d(
Function=(fnLeq),
LegendText="[L]",
Color = RGB::Blue,
XMin=(LR_Ratio_min),
XMax=(LR_Ratio_max),
XName=(LR_Ratio),
TitlePositionX=(0),
LineWidth=LW):
pReq:= plot::Function2d(
Function=(fnReq),
LegendText="[R]",
Color = RGB::Black,
XMin=(LR_Ratio_min),
XMax=(LR_Ratio_max),
XName=(LR_Ratio),
TitlePositionX=(0),
LineWidth=LW):
pR2L2eq:= plot::Function2d(
Function=(fnR2L2eq),
LegendText="[R2L2]",
Color = RGB::Red,
XMin=(LR_Ratio_min),
XMax=(LR_Ratio_max),
XName=(LR_Ratio),
TitlePositionX=(0),
LineWidth=LW):
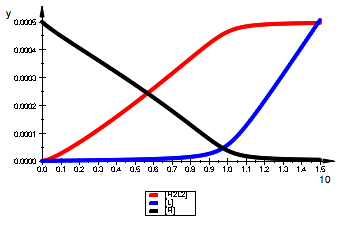
plot(pR2L2eq,pLeq,pReq,LegendVisible=TRUE);
![]()
![]()

Leq , 1.960592105e-21, 0.0005063940189
Req , 0.0004999999992, 0.000004397139854
RLeq , 1.960592105e-21, 0.00000002226685322
R2eq , 0.0002499999992, 0.0000000193348389
R2Leq , 9.802960493e-20, 0.000001958209355
R2L2eq , 9.609803474e-35, 0.0004958127525
6. Save results on disk for future use
(you can retrieve them later by executing: fread(filename,Quiet))
ProjectName;
CurrentPath
![]()
![]()
Save all numeric solutions:
filename:=CurrentPath.ProjectName.".mb";
write(filename,
// save basic equations
fRtot_B_R2_R2L2,
Req_B_R2_R2L2,
R2eq_B_R2_R2L2,
RLeq_B_R2_R2L2,
R2Leq_B_R2_R2L2,
R2L2eq_B_R2_R2L2,
// save dependent constants
KB2_B_R2_R2L2,
KB3_B_R2_R2L2,
// constants in terms of microscopic constants
KA2_B_R2_R2L2,
KB2_B_R2_R2L2,
KA3_B_R2_R2L2,
// save numeric procedures
pnLeq_B_R2_R2L2,
pnReq_B_R2_R2L2,
pnR2eq_B_R2_R2L2,
pnRLeq_B_R2_R2L2,
pnR2Leq_B_R2_R2L2,
pnR2L2eq_B_R2_R2L2
)
![]()
Relationships between kinetic constants
Conclusions
1. This system is analytically insoluble.
2. I derived numeric solutions in MACROSCOPIC constants and saved them.
3. Line shape and any kinetic simulations must be done with microscopic constants.