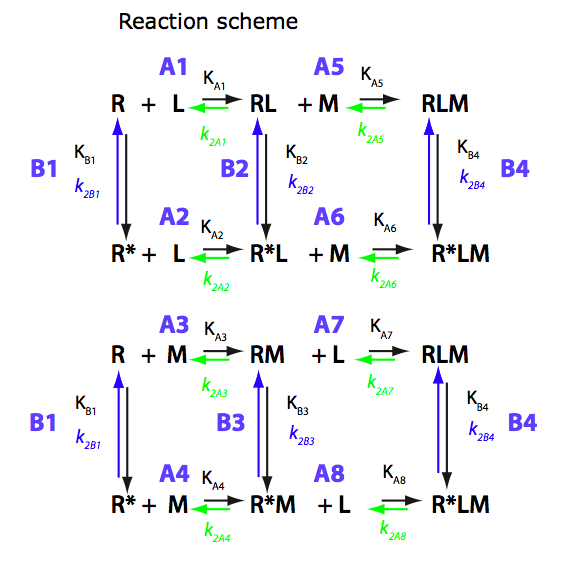
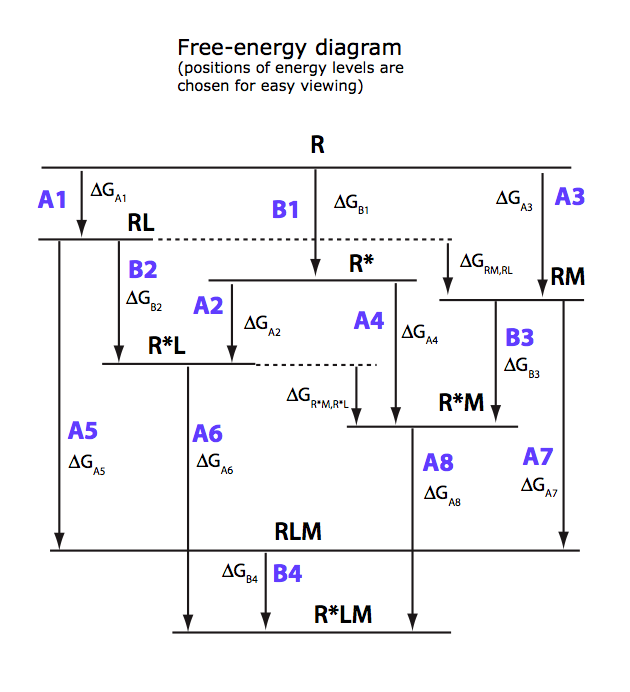
U-R-RL-RM-RLM
Binding of two ligands at non-mutually exclusive binding sites coupled with intramolecular isomerization of the receptor
(allosterically linked binding sites).

(images from
Equilibrium_models/U-R-RL-RM_RLM_model.pdf)
2. Basic equilibrium equations
3. Derivation of equations for equilibrium concentrations
4. Prepare equations for a numeric solution
5. Save results on disk for future use
In this notebook I will derive equations for two-ligand U-R-RL-RM-RLM model with separate (non-exclusive) binding sites and prepare equations for numerical simulations.
Analysis will be done in
EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/LRIM_U_R_RL_RM_RLM_analysis.mn
clean up workspace
reset()
Set path to save results into:
ProjectName:="U_R_RL_RM_RLM";
CurrentPath:="/Users/kovrigin/Documents/Workspace/Data/Data.XV/EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/";
![]()
![]()
Binding and isomerization constants
All binding constants I am using are association constants.
These relationships serve as restraints for solve(), but not restrict these values in calculations!
K_A_1
K_A_1 ;
assumeAlso(K_A_1 > 0):
assumeAlso(K_A_1 , R_):
![]()
K_A_2
K_A_2 ;
assumeAlso(K_A_2 > 0):
assumeAlso(K_A_2 , R_):
![]()
K_A_3
K_A_3 ;
assumeAlso(K_A_3 > 0):
assumeAlso(K_A_3 , R_):
![]()
K_A_4
K_A_4 ;
assumeAlso(K_A_4 > 0):
assumeAlso(K_A_4 , R_):
![]()
K_A_5
K_A_5 ;
assumeAlso(K_A_5 > 0):
assumeAlso(K_A_5 , R_):
![]()
K_A_6
K_A_6 ;
assumeAlso(K_A_6 > 0):
assumeAlso(K_A_6 , R_):
![]()
K_A_7
K_A_7 ;
assumeAlso(K_A_7 > 0):
assumeAlso(K_A_7 , R_):
![]()
K_A_8
K_A_8 ;
assumeAlso(K_A_8 > 0):
assumeAlso(K_A_8 , R_):
![]()
Isomerization constants are formation constants for the alternative species
K_B_1
K_B_1 ;
assumeAlso(K_B_1 > 0):
assumeAlso(K_B_1 , R_):
![]()
K_B_2
K_B_2 ;
assumeAlso(K_B_2 > 0):
assumeAlso(K_B_2 , R_):
![]()
K_B_3
K_B_3 ;
assumeAlso(K_B_3 > 0):
assumeAlso(K_B_3 , R_):
![]()
K_B_4
K_B_4 ;
assumeAlso(K_B_4 > 0):
assumeAlso(K_B_4 , R_):
![]()
Total concentrations
Rtot - total concentration of the receptor
Rtot;
assumeAlso(Rtot>0):
assumeAlso(Rtot,R_):
![]()
Ltot - total concentration of the first ligand
Ltot;
assumeAlso(Ltot>0):
assumeAlso(Ltot,R_):
![]()
Mtot - total concentration of the second ligand
Mtot;
assumeAlso(Mtot>0):
assumeAlso(Mtot,R_):
![]()
Common equilibrium concentrations
Req - equilibrium concentration of a receptor monomer
Req;
assumeAlso(Req>0):
assumeAlso(Req<Rtot):
assumeAlso(Req,R_):
![]()
Rstareq - equilibrium concentration of an isomerized receptor monomer
Rstareq;
assumeAlso(Rstareq>0):
assumeAlso(Rstareq<Rtot):
assumeAlso(Rstareq,R_):
![]()
Leq - equilibrium concentration of a free ligand
Leq;
assumeAlso(Leq>0):
assumeAlso(Leq<Ltot):
assumeAlso(Leq,R_):
![]()
RLeq - equilibrium concentration of a receptor-ligand complex
RLeq;
assumeAlso(RLeq>0):
assumeAlso(RLeq<Rtot):
assumeAlso(RLeq,R_):
![]()
RstarLeq - equilibrium concentration of an isomerized receptor-ligand complex
RstarLeq;
assumeAlso(RstarLeq>0):
assumeAlso(RstarLeq<Rtot):
assumeAlso(RstarLeq,R_):
![]()
RMeq - equilibrium concentration of a receptor-ligand complex
RMeq;
assumeAlso(RMeq>0):
assumeAlso(RMeq<Rtot):
assumeAlso(RMeq,R_):
![]()
RstarMeq - equilibrium concentration of a receptor-ligand complex
RstarMeq;
assumeAlso(RstarMeq>0):
assumeAlso(RstarMeq<Rtot):
assumeAlso(RstarMeq,R_):
![]()
RLMeq - equilibrium concentration of a receptor-double-ligand complex
RLMeq;
assumeAlso(RLMeq>0):
assumeAlso(RLMeq<Rtot):
assumeAlso(RLMeq,R_):
![]()
RstarLMeq - equilibrium concentration of a receptor-ligand complex
RstarLMeq;
assumeAlso(RstarLMeq>0):
assumeAlso(RstarLMeq<Rtot):
assumeAlso(RstarLMeq,R_):
![]()
anames(Properties,User);

2. Basic equilibrium equations
I will express Rtot as a function of Leq and all constants for numeric solution
Total concentrations of a receptor and a ligand
eq2_1:= Rtot = Req + Rstareq + RLeq + RstarLeq + RMeq + RstarMeq + RLMeq + RstarLMeq;
eq2_2:= Ltot = Leq + RLeq + RstarLeq + RLMeq + RstarLMeq;
eq2_3:= Mtot = Meq + RMeq + RstarMeq + RLMeq + RstarLMeq;
![]()
![]()
![]()
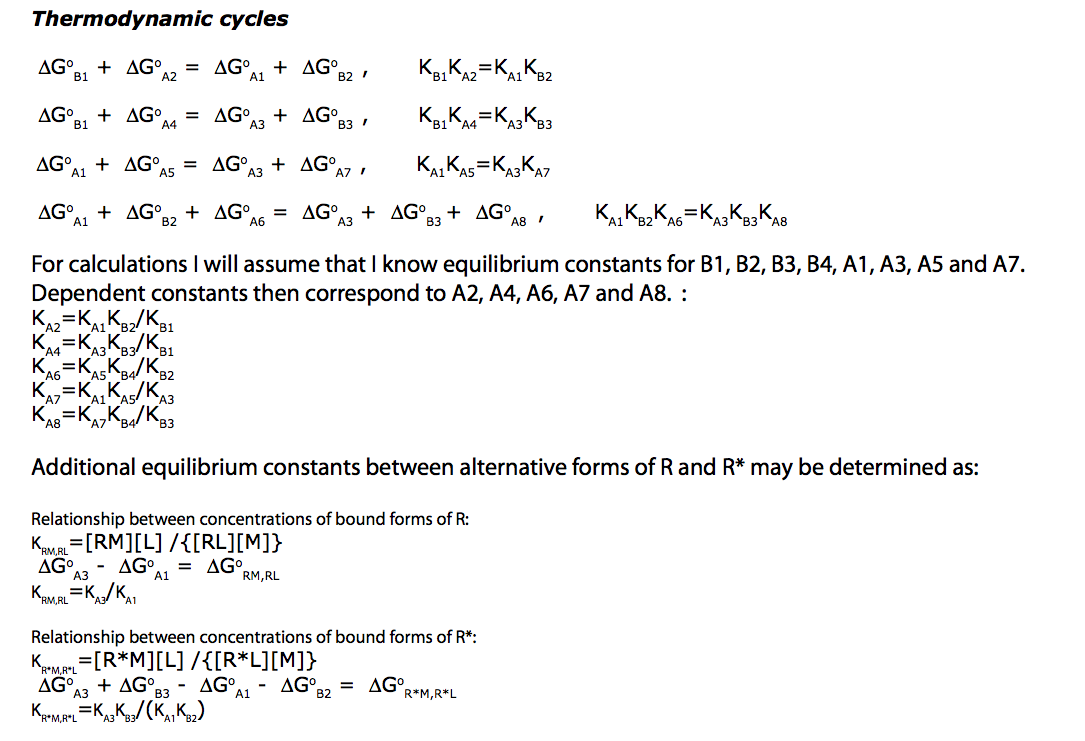
Write equilibrium thermodynamics equations (KA2 and KA4 are chosen to be dependent constants).
eq2_4:= K_A_1 = RLeq / (Req*Leq)
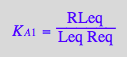
eq2_5:= K_A_2 = RstarLeq / (Rstareq*Leq) // !!! Dependent constant
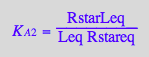
eq2_6:= K_A_3 = RMeq / (Req*Meq)
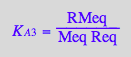
eq2_7:= K_A_4 = RstarMeq / (Rstareq*Meq) // !!! Dependent constant
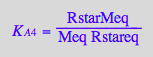
eq2_8:= K_A_5 = RLMeq / (RLeq*Meq)
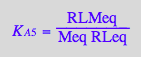
eq2_9:= K_A_6 = RstarLMeq / (RstarLeq*Meq) // !!! Dependent constant
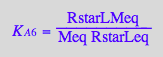
eq2_10:= K_A_7 = RLMeq /(RMeq*Leq)
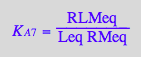
eq2_11:= K_A_8 = RstarLMeq/(RstarMeq*Leq) // !!! Dependent constant

eq2_12:= K_B_1 = Rstareq / Req
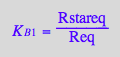
eq2_13:= K_B_2 = RstarLeq / RLeq
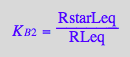
eq2_14:= K_B_3 = RstarMeq / RMeq

eq2_15:= K_B_4 = RstarLMeq / RLMeq

3. Derivation of equations for equilibrium concentrations
Try to express Leq as a function of all constants and total concentrations
Express RstarLMeq
eq2_15;
solve(%,RstarLMeq);
%[1][1];
eq3_1:= RstarLMeq=%

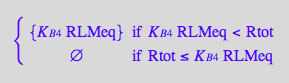
![]()
![]()
Express RstarMeq
eq2_14;
solve(%,RstarMeq);
%[1][1];
eq3_2:= RstarMeq=%


![]()
![]()
Express RstarLeq
eq2_13;
solve(eq2_13,RstarLeq);
%[1][1];
eq3_3:= RstarLeq = %
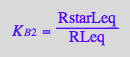
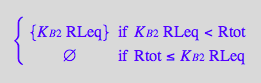
![]()
![]()
Express Rstareq
eq2_12;
solve(eq2_12,Rstareq);
%[1][1];
eq3_4:= Rstareq = %
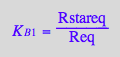

![]()
![]()
Express RLMeq
eq2_10;
solve(eq2_10,RLMeq);
%[1][1];
eq3_5:= RLMeq = %
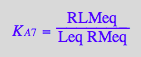
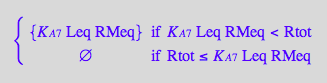
![]()
![]()
Express RMeq
eq2_6;
solve(eq2_6,RMeq);
%[1][1];
eq3_6:= RMeq = %
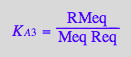
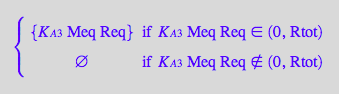
![]()
![]()
Express RLeq
eq2_4;
solve(eq2_4,RLeq);
%[1][1];
eq3_7:= RLeq = %
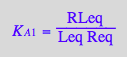

![]()
![]()
Substitute in mass conservation equations (begin with shorter ones for ligands)
eq2_2;
% | eq3_1;
% | eq3_3;
% | eq3_5;
% | eq3_6;
% | eq3_7;
eq3_8:= %:
![]()
![]()
![]()
![]()
![]()
![]()
Express [R] from there (keep [L] for last solving)
solve(eq3_8, Req);
%[1][1];
eq3_9:= Req = %
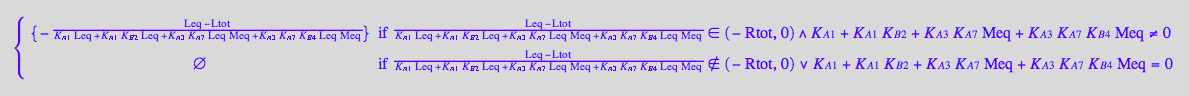
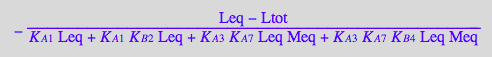
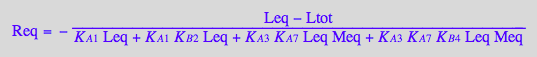
Use second ligand mass conservation law
eq2_3;
% | eq3_1;
% | eq3_2;
% | eq3_5;
% | eq3_6;
% | eq3_9;
eq3_10:= %:
![]()
![]()
![]()
![]()
![]()

Solve it for Meq
solutions3_10:=solve(eq3_10,Meq):
eq3_11:= solutions3_10[i,1] $ i=1..nops(solutions3_10):
nops(%)
![]()
How many independent soltions we have?
Is 1st solution a combination of 2nd and 3rd?
solution1:=eq3_11[4]: // a sequence of roots
solution2:=eq3_11[2][1]: // extract equation out of a sequence
solution3:=eq3_11[3][1]: // extract equation out of a sequence
if solution2 in solution1
then print(Unquoted,"First set of roots contains the second root.");
else print(Unquoted,"First set of roots DOES NOT contain the second root!");
end_if;
if solution3 in solution1
then print(Unquoted,"First set of roots contains the third root.");
else print(Unquoted,"First set of roots DOES NOT contain the third root!");
end_if;
First set of roots contains the second root.
First set of roots contains the third root.
Check correctness of the solutions by substitution into original equation solved:
Check first root
test1:= eq3_10 | Meq=solution2:
normal(%);
![]()
Check first root
test2:= eq3_10 | Meq=solution3:
normal(%);
![]()
Both solutions are correct.
Choose meaningful one for further work
solution2 | Leq=1 | Rtot=1 | Mtot=1 | Ltot=1 | K_A_1 =1 | K_A_3 =1 | K_A_5 =1 | K_A_7 =1 | K_A_8 =1 | K_B_1 =1 | K_B_2 =1 | K_B_3 =1 | K_B_4 =1 :
float(%)
![]()
solution3 | Leq=1 | Rtot=1 | Mtot=1 | Ltot=1 | K_A_1 =1 | K_A_3 =1 | K_A_5 =1 | K_A_7 =1 | K_A_8 =1 | K_B_1 =1 | K_B_2 =1 | K_B_3 =1 | K_B_4 =1 :
float(%)
![]()
Solution 3 is meaningful !
ASSIGN TO AN EQUATION:
eq3_12:= Meq = solution3:
NOTE: Here I did not pay attention, but A5 and A7 are dependent. Current equations are cast in A7. I will change it to A5 using relationship:
eq3_14:= K_A_7=K_A_1*K_A_5/K_A_3
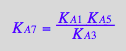
Finally, substitute all into a mass conservation law for a receptor (40 seconds to display!)
eq2_1;
% | eq3_1 ;
% | eq3_2 ;
% | eq3_3 ;
% | eq3_4 ;
% | eq3_5 ;
% | eq3_6 ;
% | eq3_7 ;
% | eq3_9 ;
% | eq3_12 :
% | eq3_14 :
eq3_13:=%:
![]()
![]()
![]()
![]()
![]()
![]()
![]()
![]()
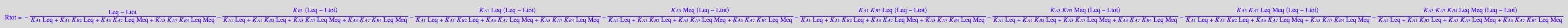
Summary of final equations
Reassign names for storage
ProjectName
![]()
Rtot=f([L])
Final equation (40 seconds to display!)
Rtot_U_R_RL_RM_RLM:=eq3_13:
[M]
Meq_U_R_RL_RM_RLM:=eq3_12 | eq3_14

[R]
Req_U_R_RL_RM_RLM:=eq3_9 | eq3_14

[RL]
RLeq_U_R_RL_RM_RLM:=eq3_7
![]()
[RM]
RMeq_U_R_RL_RM_RLM:=eq3_6
![]()
[RLM]
RLMeq_U_R_RL_RM_RLM:=eq3_5 | eq3_14

[R*]
Rstareq_U_R_RL_RM_RLM:=eq3_4
![]()
[R*L]
RstarLeq_U_R_RL_RM_RLM:=eq3_3
![]()
[R*M]
RstarMeq_U_R_RL_RM_RLM:=eq3_2
![]()
[R*LM]
RstarLMeq_U_R_RL_RM_RLM:=eq3_1
![]()
Test equations' consistency by expressing dependentt equilibrium constants:
eq2_5;
eq3_15:= KA2_U_R_RL_RM_RLM:= % | eq3_1 | eq3_2 | eq3_3 | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_9 | eq3_12 | eq3_13 ;
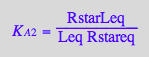
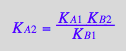
Correct !
eq2_7;
eq3_16:= KA4_U_R_RL_RM_RLM:= % | eq3_1 | eq3_2 | eq3_3 | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_9 | eq3_12 | eq3_13 ;
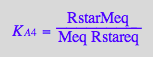
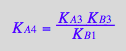
correct!
eq2_9;
eq3_17:= KA6_U_R_RL_RM_RLM:= % | eq3_1 | eq3_2 | eq3_3 | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_9 | eq3_12 | eq3_13 ;
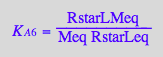
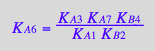
correct
eq2_11;
eq3_18:= KA8_U_R_RL_RM_RLM:= % | eq3_1 | eq3_2 | eq3_3 | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_9 | eq3_12 | eq3_13 ;

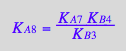
correct !
5. Save results on disk for future use
(you can retrieve them later by executing: fread(filename,Quiet))
ProjectName
![]()
filename:=CurrentPath.ProjectName.".mb";
write(filename,
Rtot_U_R_RL_RM_RLM,
Meq_U_R_RL_RM_RLM,
Req_U_R_RL_RM_RLM,
RMeq_U_R_RL_RM_RLM,
RLeq_U_R_RL_RM_RLM,
Rstareq_U_R_RL_RM_RLM,
RstarMeq_U_R_RL_RM_RLM,
RstarLeq_U_R_RL_RM_RLM,
RLMeq_U_R_RL_RM_RLM,
RstarLMeq_U_R_RL_RM_RLM,
KA2_U_R_RL_RM_RLM,
KA4_U_R_RL_RM_RLM,
KA6_U_R_RL_RM_RLM,
KA8_U_R_RL_RM_RLM)
![]()