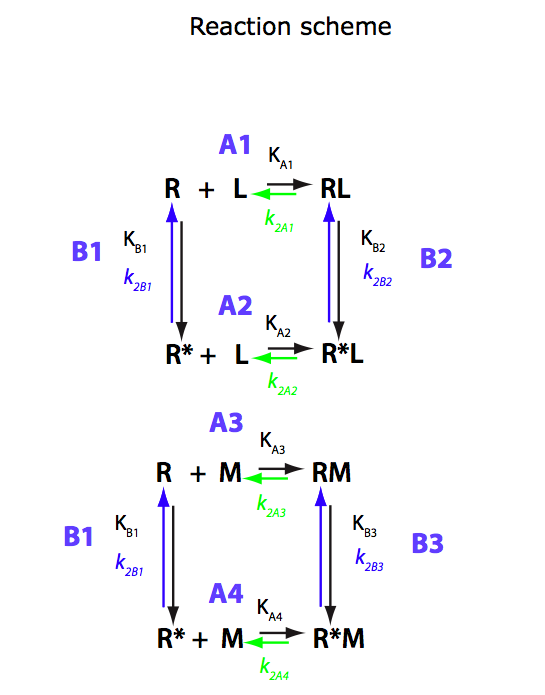
U-R-RL-RM
Binding of two mutually exclusive ligands coupled with intramolecular isomerization of the receptor (competitive ligand binding)

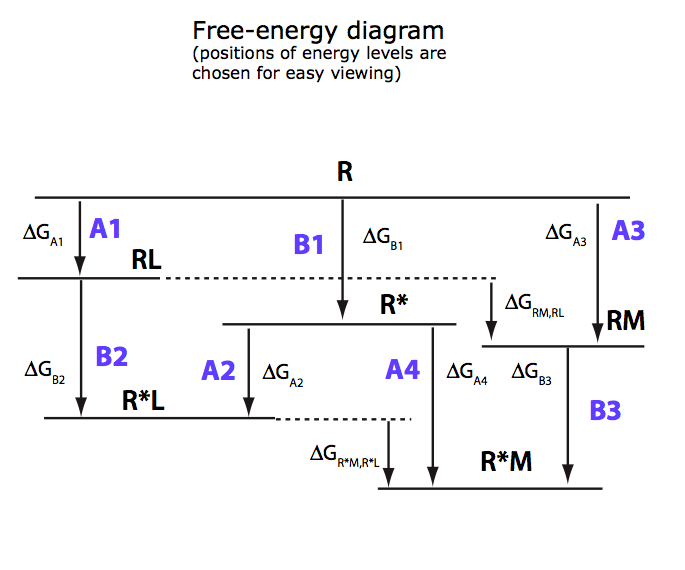
(images from Equilibrium_models/U-R-RL-RM_model.pdf)
2. Basic equilibrium equations
3. Derivation of equations for equilibrium concentrations
4. Prepare equations for a numeric solution
6. Preparing numeric solutions
7. Reproduce a graph for equilibrium concentrations using a numeric solution
8. Save results on disk for future use
In this notebook I will derive equations for two-ligand competitive U-R-RL-RM model and prepare equations for numerical simulations.
Analysis will be done in
EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/LRIM_U_R_RL_RM_analysis.mn
clean up workspace
reset()
Set path to save results into:
ProjectName:="U_R_RL_RM";
CurrentPath:="/Users/kovrigin/Documents/Workspace/Data/Data.XV/EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/";
![]()
![]()
Binding and isomerization constants
All binding constants I am using are association constants.
These relationships serve as restraints for solve(), but not restrict these values in calculations!
K_A_1
K_A_1 ;
assumeAlso(K_A_1 > 0):
assumeAlso(K_A_1 , R_):
![]()
K_A_2
K_A_2 ;
assumeAlso(K_A_2 > 0):
assumeAlso(K_A_2 , R_):
![]()
K_A_3
K_A_3 ;
assumeAlso(K_A_3 > 0):
assumeAlso(K_A_3 , R_):
![]()
K_A_4
K_A_4 ;
assumeAlso(K_A_4 > 0):
assumeAlso(K_A_4 , R_):
![]()
Isomerization constants are formation constants for the alternative species
K_B_1
K_B_1 ;
assumeAlso(K_B_1 > 0):
assumeAlso(K_B_1 , R_):
![]()
K_B_2
K_B_2 ;
assumeAlso(K_B_2 > 0):
assumeAlso(K_B_2 , R_):
![]()
K_B_3
K_B_3 ;
assumeAlso(K_B_3 > 0):
assumeAlso(K_B_3 , R_):
![]()
Total concentrations
Rtot - total concentration of the receptor
Rtot;
assumeAlso(Rtot>0):
assumeAlso(Rtot,R_):
![]()
Ltot - total concentration of a ligand
Ltot;
assumeAlso(Ltot>0):
assumeAlso(Ltot,R_):
![]()
Common equilibrium concentrations
Req - equilibrium concentration of a receptor monomer
Req;
assumeAlso(Req>0):
assumeAlso(Req<Rtot):
assumeAlso(Req,R_):
![]()
Rstareq - equilibrium concentration of an isomerized receptor monomer
Rstareq;
assumeAlso(Rstareq>0):
assumeAlso(Rstareq<Rtot):
assumeAlso(Rstareq,R_):
![]()
Leq - equilibrium concentration of a free ligand
Leq;
assumeAlso(Leq>0):
assumeAlso(Leq<Ltot):
assumeAlso(Leq,R_):
![]()
RLeq - equilibrium concentration of a receptor-ligand complex
RLeq;
assumeAlso(RLeq>0):
assumeAlso(RLeq<Rtot):
assumeAlso(RLeq,R_):
![]()
RstarLeq - equilibrium concentration of an isomerized receptor-ligand complex
RstarLeq;
assumeAlso(RstarLeq>0):
assumeAlso(RstarLeq<Rtot):
assumeAlso(RstarLeq,R_):
![]()
RMeq - equilibrium concentration of a receptor-ligand complex
RMeq;
assumeAlso(RMeq>0):
assumeAlso(RMeq<Rtot):
assumeAlso(RMeq,R_):
![]()
RstarMeq - equilibrium concentration of a receptor-ligand complex
RstarMeq;
assumeAlso(RstarMeq>0):
assumeAlso(RstarMeq<Rtot):
assumeAlso(RstarMeq,R_):
![]()
anames(Properties,User);
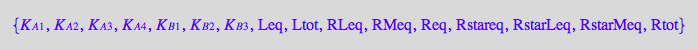
2. Basic equilibrium equations
Working equation: I will try to express analytical [L] from equation for a total concentration of a receptor or use the expression for a numeric solution if analytical is not possible
Total concentrations of a receptor and a ligand
eq2_1:= Rtot = Req + Rstareq + RLeq + RstarLeq + RMeq + RstarMeq;
eq2_2:= Ltot = Leq + RLeq + RstarLeq;
eq2_3:= Mtot = Meq + RMeq + RstarMeq;
![]()
![]()
![]()
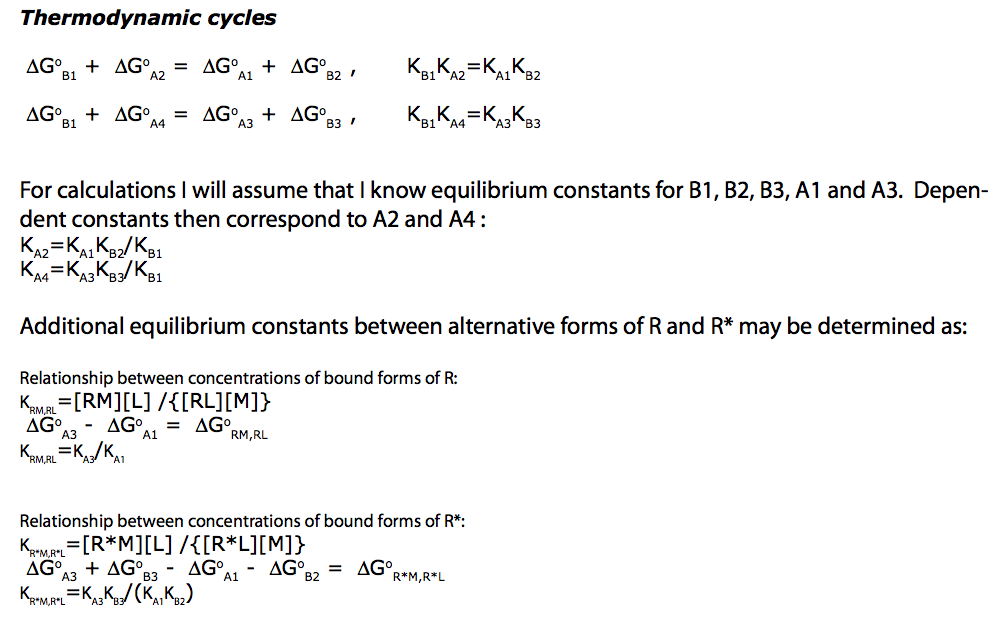
Write equilibrium thermodynamics equations (KA2 and KA4 are chosen to be dependent constants).
eq2_4:= K_A_1 = RLeq / (Req*Leq)
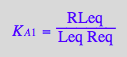
eq2_5:= K_A_2 = RstarLeq / (Rstareq*Leq) // !!! Dependent constant
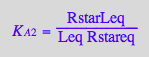
eq2_6:= K_A_3 = RMeq / (Req*Meq)
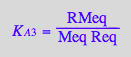
eq2_7:= K_A_4 = RstarMeq / (Rstareq*Meq) // !!! Dependent constant
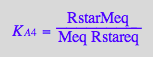
eq2_8:= K_B_1 = Rstareq / Req
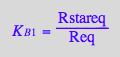
eq2_9:= K_B_2 = RstarLeq / RLeq
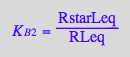
eq2_10:= K_B_3 = RstarMeq / RMeq

3. Derivation of equations for equilibrium concentrations
Try to express Leq as a function of all constants and total concentrations
Express [R*L]
eq2_9;
solve(%, RstarLeq);
%[1][1];
eq3_1:= RstarLeq = %
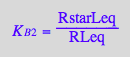
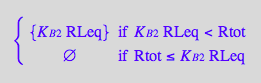
![]()
![]()
Express [R*M]
eq2_10;
solve(%, RstarMeq);
%[1][1];
eq3_2:= RstarMeq = %


![]()
![]()
Express [R*]
eq2_8;
solve(%,Rstareq);
%[1][1];
eq3_3:= Rstareq = %
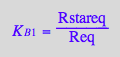

![]()
![]()
Express [RLeq]
eq2_4;
solve(%,RLeq);
%[1][1];
eq3_4:= RLeq = %
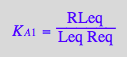

![]()
![]()
Express [RMeq]
eq2_6;
solve(%,RMeq);
%[1][1];
eq3_5:= RMeq = %
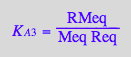
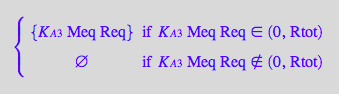
![]()
![]()
Substitute in mass conservation equations (begin with shorter ones for ligands)
eq2_2;
% | eq3_1;
% | eq3_4;
eq3_6:= %:
![]()
![]()
![]()
Express [R] from there (keep [L] for last solving)
solve(eq3_6,Req);
%[1][1];
eq3_7:= Req = %
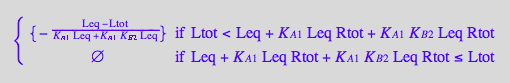
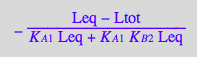
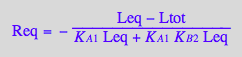
Use second ligand mass conservation law
eq2_3;
% | eq3_2;
% | eq3_5;
% | eq3_7;
eq3_8:= %:
![]()
![]()
![]()
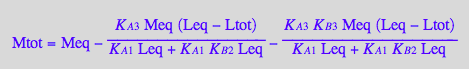
Solve it for [M]
solution3_8:=solve(eq3_8,Meq);
%[1][1];
eq3_9:= Meq = %

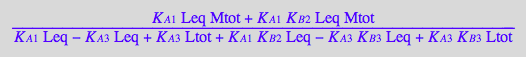
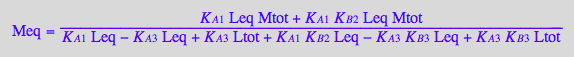
Use receptor mass conservation law
eq2_1;
% | eq3_2;
% | eq3_1;
% | eq3_3;
% | eq3_4;
% | eq3_5;
% | eq3_9;
% | eq3_7;
eq3_10:=%:
![]()
![]()
![]()
![]()
![]()
![]()
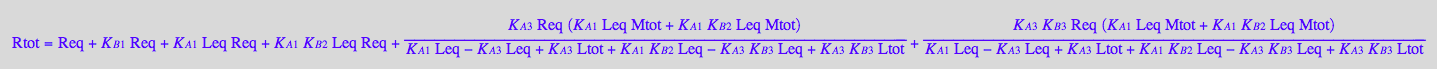

Solve for [L]
solution3_10:= solve(eq3_10,Leq);
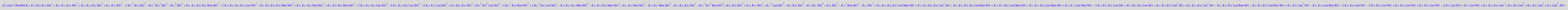
Insoluble in analytical form.
4. Prepare equations for a numeric solution
I will express all equilibrium concentrations in terms of each other in successive order, easy to code in Matlab.
Summary of equilibrium thermodynamics equations for U-R-RL-RM system
[L] (to be solved for)
eq3_10

[M]
eq3_9
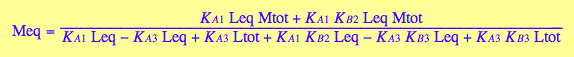
[R]
eq3_7
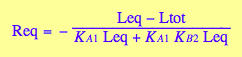
[RM]
eq3_5
![]()
[RL]
eq3_4;
![]()
[R*]
eq3_3;
![]()
[R*M]
eq3_2;
![]()
[R*L]
eq3_1
![]()
Test equations' consistency by expressing dependentt equilibrium constants:
eq2_7;
% | eq3_3;
% | eq3_2;
% | eq3_5;
eq3_11:=%:
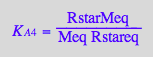

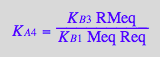
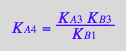
Correct relationship!
eq2_5;
% | eq3_3;
% | eq3_1;
% | eq3_4;
eq3_12:=%:
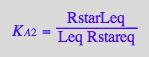

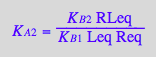
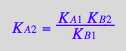
Correct relationship!
6. Preparing numeric solutions
Make a function for numeric solving and switch to LRratio=Ltot/Rtot
fRtot_U_R_RL_RM:=(Rtot, LR_Ratio, MR_Ratio, Leq, K_A_1, K_A_3, K_B_1, K_B_2, K_B_3) --> eq3_10[2] | Ltot=Rtot*LR_Ratio | Mtot=Rtot*MR_Ratio

Assume some constant values for testing
Total_R:=1e-3:
Ka1:=1e7:
Ka3:=1e6:
Kb1:=2/1:
Kb2:=0.1:
Kb3:=0.1:
LRratio_max:=1.2:
MRratio_max:=1.2:
// some test point
LRratio:=0.95:
MRratio:=0.95:
Define a procedure for numeric solving the equation Rtot=f([L]) for [L] thus creating a function [L]=f(Rtot,...)
// WARNING: make sure the Leq search range starts with a non-zero number!!!! Use a number larger than that to create approximation of Ltot=0
pnLeq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
begin
// numeric solving equation for Leq in a restricted range.
// WARNING: make sure the range starts with a non-zero number!!!!
result:=numeric::fsolve(
Total_R-fRtot_U_R_RL_RM(Total_R, LRratio, MRratio, Leq, Ka1, Ka3, Kb1, Kb2, Kb3),
Leq=10e-32..Total_R*LRratio);
//print(result);
// extract answer from equation
result[1][2]
end_proc;
![]()
test operation
pnLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
Make "portable" equations (unique names for future use):
Meq_U_R_RL_RM:=eq3_9;
Req_U_R_RL_RM:=eq3_7;
RMeq_U_R_RL_RM:=eq3_5;
RLeq_U_R_RL_RM:=eq3_4;
Rstareq_U_R_RL_RM:=eq3_3;
RstarMeq_U_R_RL_RM:=eq3_2;
RstarLeq_U_R_RL_RM:=eq3_1;
KA2_U_R_RL_RM:=eq3_12;
KA4_U_R_RL_RM:=eq3_11;
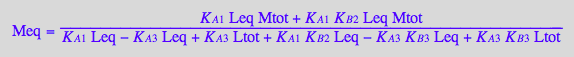
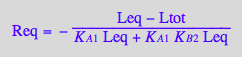
![]()
![]()
![]()
![]()
![]()
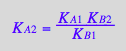
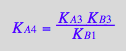
Define functions for equilibrium concentrations of all species:
[R]
Req_U_R_RL_RM;
pnReq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local L;
begin
L:=pnLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [R]
Req_U_R_RL_RM[2] | Ltot=Total_R*LRratio | Leq=L | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
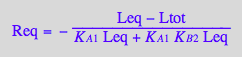
![]()
Test
pnReq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
[M]
Meq_U_R_RL_RM;
pnMeq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local L;
begin
L:=pnLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [M]
Meq_U_R_RL_RM[2] | Ltot=Total_R*LRratio | Mtot=Total_R*MRratio | Leq=L | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
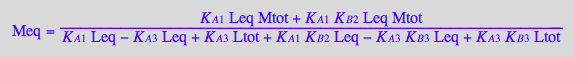
![]()
Test
pnMeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
[RL]
RLeq_U_R_RL_RM;
pnRLeq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local R, L;
begin
L:=pnLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
R:=pnReq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [RL]
RLeq_U_R_RL_RM[2] | Req=R | Leq=L | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
![]()
![]()
Test
pnRLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
[RM]
RMeq_U_R_RL_RM;
pnRMeq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local M, R;
begin
R:=pnReq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
M:=pnMeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [RM]
RMeq_U_R_RL_RM[2] | Req=R | Meq=M | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
![]()
![]()
Test
pnRMeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
[R*]
Rstareq_U_R_RL_RM;
pnRstareq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local R;
begin
R:=pnReq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [R*]
Rstareq_U_R_RL_RM[2] | Req=R | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
![]()
![]()
Test
pnRstareq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
[R*L]
RstarLeq_U_R_RL_RM;
pnRstarLeq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local RL;
begin
RL:=pnRLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [R*L]
RstarLeq_U_R_RL_RM[2] | RLeq=RL | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
![]()
![]()
Test
pnRstarLeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
[R*M]
RstarMeq_U_R_RL_RM;
pnRstarMeq_U_R_RL_RM:= proc(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
// Parameter names should be different from
// variable names used in the equation!!!
local RM;
begin
RM:=pnRMeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3);
// Equation for [R*M]
RstarMeq_U_R_RL_RM[2] | RMeq=RM | K_A_1=Ka1 | K_A_3=Ka3 | K_B_1=Kb1 | K_B_2=Kb2 | K_B_3=Kb3;
end_proc
![]()
![]()
Test
pnRstarMeq_U_R_RL_RM(Total_R, LRratio, MRratio, Ka1, Ka3, Kb1, Kb2, Kb3)
![]()
8. Save results on disk for future use
(you can retrieve them later by executing: fread(filename,Quiet))
ProjectName
![]()
Save all numeric solutions:
filename:=CurrentPath.ProjectName.".mb";
write(filename,
// save basic equations
Meq_U_R_RL_RM,
Req_U_R_RL_RM,
RMeq_U_R_RL_RM,
RLeq_U_R_RL_RM,
Rstareq_U_R_RL_RM,
RstarMeq_U_R_RL_RM,
RstarLeq_U_R_RL_RM,
// save dependent constants
KA2_U_R_RL_RM,
KA4_U_R_RL_RM,
// save numeric procedures
fRtot_U_R_RL_RM,
pnLeq_U_R_RL_RM,
pnMeq_U_R_RL_RM,
pnReq_U_R_RL_RM,
pnRMeq_U_R_RL_RM,
pnRLeq_U_R_RL_RM,
pnRstareq_U_R_RL_RM,
pnRstarMeq_U_R_RL_RM,
pnRstarLeq_U_R_RL_RM)
![]()
Conclusions
1. This system is analytically insoluble.
2. I derived numeric solutions and saved them.