Collection of simulations for
U-R-RL-RM
Numerical simulation is done in
Equilibria/U-R-RL-RM_analysis.mn
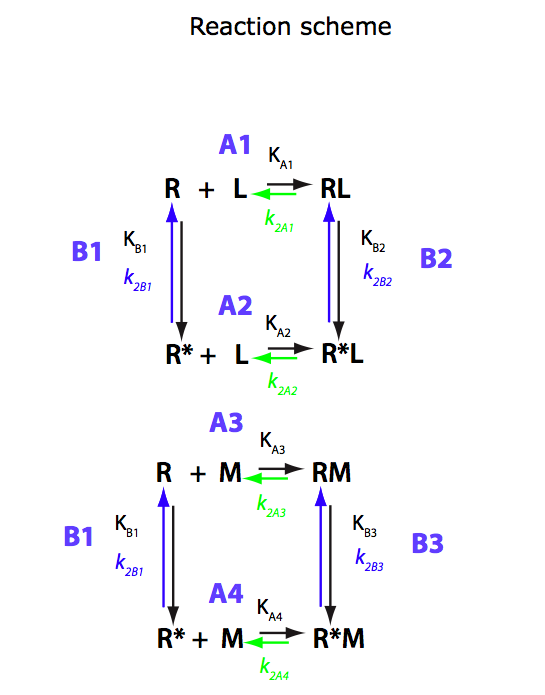
Binding of two mutually exclusive ligands coupled with intramolecular isomerization of the receptor (competitive ligand binding)

Simple competitive binding to the same site without any isomerization
Isomerization produces forms strongly favored by different ligands
In this notebook I will collect results of simulations done for U-R-RL-RM model in
Equilibria/U-R-RL-RM_analysis.mn
Simple competitive binding to the same site without any isomerization
L has 100-fold higher affinity than M
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=100000000.0: //Ka2:=100000000.0(dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=100000000.0: //Ka2:=100000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=100000000.0: //Ka2:=100000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=100000000.0: //Ka2:=100000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
L has 10-fold higher affinity than M
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=10000000.0: //Ka2:=10000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=10000000.0: //Ka2:=10000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=10000000.0: //Ka2:=10000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=10000000.0: //Ka2:=10000000.0 (dep.) Ka3:=1000000.0: //Ka4:=1000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
Affinities of L and M to R are equal:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
L has 10-fold lower affinity than M
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=10000000.0: //Ka4:=10000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=10000000.0: //Ka4:=10000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=10000000.0: //Ka4:=10000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=10000000.0: //Ka4:=10000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
L has 100-fold lower affinity than M
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=100000000.0: //Ka4:=100000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=100000000.0: //Ka4:=100000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=100000000.0: //Ka4:=100000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=1000000.0: //Ka2:=1000000.0 (dep.) Ka3:=100000000.0: //Ka4:=100000000.0 (dep.) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
Conclusion:
1. Main observation: Before reaching equivalence point at L/R=M/R titration proceeds like only utilizing free R form only.
2. After this point a new non-linear segment of the RL curve appears where displacement of M by L occurs with apparent low affinity binding behavior.
Isomerization produces forms strongly favored by different ligands
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=1000000.0: //Ka2:=500.0 (dep.) Ka3:=1000.0: //Ka4:=500000.0 (dep.) Kb1:=2: Kb2:=0.001: Kb3:=1000.0:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=1000000.0: //Ka2:=500.0 (dep.) Ka3:=1000.0: //Ka4:=500000.0 (dep.) Kb1:=2: Kb2:=0.001: Kb3:=1000.0:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=1000000.0: //Ka2:=500.0 (dep.) Ka3:=1000.0: //Ka4:=500000.0 (dep.) Kb1:=2: Kb2:=0.001: Kb3:=1000.0:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=1000000.0: //Ka2:=500.0 (dep.) Ka3:=1000.0: //Ka4:=500000.0 (dep.) Kb1:=2: Kb2:=0.001: Kb3:=1000.0:
|
This result is very similar to the case with no isomerization and equal affinities of L and M to R :
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1.0e-16: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.3: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=0.6: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
U_R_RL_RM, Titration with L Total_R:=0.001: MRratio:=1: Ka1:=1000000.0: //Ka2:=1000000.0 (dependent) Ka3:=1000000.0: //Ka4:=1000000.0 (dependent) Kb1:=1.0e-16: Kb2:=1.0e-16: Kb3:=1.0e-16:
|
Conclusion:
Thermodynamically, there is no difference between the case when two ligands compete for the same binding site or binding to different, mutually exclusive receptor forms. It will likely only have an equilibrium constant between the receptor froms factored into the apparent affinities.
Conclusions
(copied from sections above)
1. Main observation: Before reaching equivalence point at L/R=M/R titration proceeds like only utilizing free R form only.
3. After this point a new non-linear segment of the RL curve appears where displacement of M by L occurs with apparent low affinity binding behavior.
2. Thermodynamically, there is no difference between the case when two ligands compete for the same binding site or binding to different, mutually exclusive receptor forms. It will likely only have an equilibrium constant between the receptor froms factored into the apparent affinities.