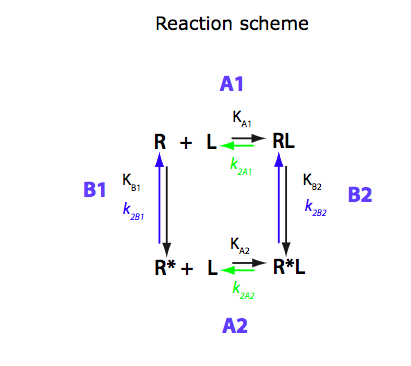
Derivation of equations for U-R-RL model
Binding of one ligand coupled with intramolecular isomerization
(this is the same model as ABC1, just in a new notation)

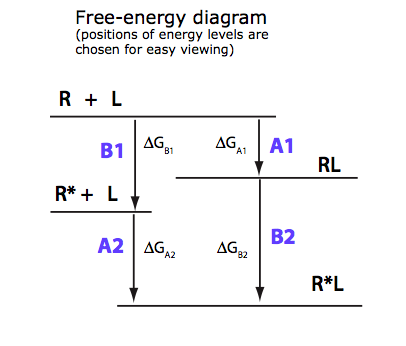
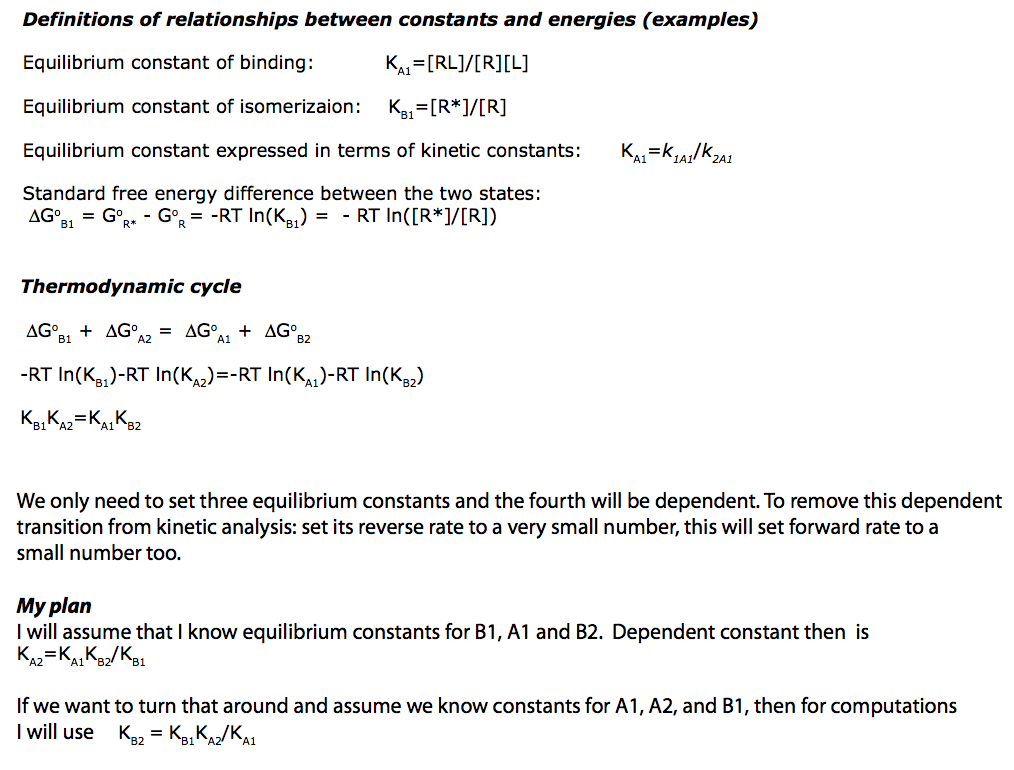
2. Derivation of a main equation
3. Define functions for equilibrium concentrations
4. Test if solution is meaningful
5. Check whether the solution satisfies all initial equation and conditions
6. Save results on disk for future use
In this notebook I will re-derive equations for U-R-RL model (previously analyzed as ABC1) and prepare equations for numerical simulations.
Analysis will be done in
EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/LRIM_U_R_RL.analysis.mn
clean up workspace
reset()
Set path to save results into:
ProjectName:="U_R_RL";
CurrentPath:="/Users/kovrigin/Documents/Workspace/Data/Data.XV/EKM16.Analysis_of_multistep_kinetic_mechanisms/Equilibria/";
![]()
![]()
Binding and isomerization constants
All binding constants I am using are association constants.
These relationships serve as restraints for solve(), but not restrict these values in calculations!
K_A_1
K_A_1 ;
assumeAlso(K_A_1 > 0):
assumeAlso(K_A_1 , R_):
![]()
K_A_2
K_A_2 ;
assumeAlso(K_A_2 > 0):
assumeAlso(K_A_2 , R_):
![]()
Isomerization constants are formation constants for the alternative species
K_B_1
K_B_1 ;
assumeAlso(K_B_1 > 0):
assumeAlso(K_B_1 , R_):
![]()
K_B_2
K_B_2 ;
assumeAlso(K_B_2 > 0):
assumeAlso(K_B_2 , R_):
![]()
Total concentrations
Rtot - total concentration of the receptor
Rtot;
assumeAlso(Rtot>0):
assumeAlso(Rtot,R_):
![]()
Ltot - total concentration of a ligand
Ltot;
assumeAlso(Ltot>0):
assumeAlso(Ltot,R_):
![]()
Common equilibrium concentrations
Req - equilibrium concentration of a receptor monomer
Req;
assumeAlso(Req>0):
assumeAlso(Req<Rtot):
assumeAlso(Req,R_):
![]()
Rstareq - equilibrium concentration of an isomerized receptor monomer
Rstareq;
assumeAlso(Rstareq>0):
assumeAlso(Rstareq<Rtot):
assumeAlso(Rstareq,R_):
![]()
Leq - equilibrium concentration of a free ligand
Leq;
assumeAlso(Leq>0):
assumeAlso(Leq<Ltot):
assumeAlso(Leq,R_):
![]()
RLeq - equilibrium concentration of a receptor-ligand complex
RLeq;
assumeAlso(RLeq>0):
assumeAlso(RLeq<Rtot):
assumeAlso(RLeq,R_):
![]()
RstarLeq - equilibrium concentration of an isomerized receptor-ligand complex
RstarLeq;
assumeAlso(RstarLeq>0):
assumeAlso(RstarLeq<Rtot):
assumeAlso(RstarLeq,R_):
![]()
anames(Properties,User);
2. Derivation of a main equation
Working equation: I will try to express analytical [L] from equation for a total concentration of a receptor or use the expression for a numeric solution if analytical is not possible
Total concentrations of a receptor and a ligand
eq2_1:= Rtot = Req + Rstareq + RLeq + RstarLeq;
eq2_2:= Ltot = Leq + RLeq + RstarLeq
![]()
![]()
Write equilibrium thermodynamics equations
eq2_3:= K_A_1 = RLeq / (Req*Leq)
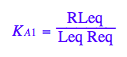
The following constant as chosen to be dependent in the model setup (don't use this equation in the solution):
eq2_4:= K_A_2 = RstarLeq / (Rstareq*Leq)
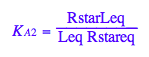
eq2_5:= K_B_1 = Rstareq/Req
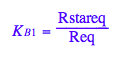
eq2_6:= K_B_2 = RstarLeq/RLeq
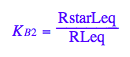
Try to express Leq as a function of all constants and total concentrations
Express RstarLeq
eq2_6;
solve(%,RstarLeq);
%[1][1];
eq2_7:= RstarLeq= %
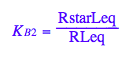
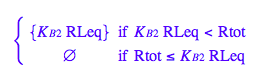
![]()
![]()
Express Rstareq
eq2_4;
% | eq2_7;
solve(%,Rstareq);
eq2_8:= Rstareq = %[1][1]
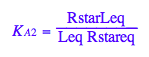
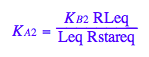

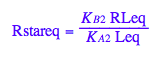
Express RLeq
eq2_5;
% | eq2_8;
solve(%,RLeq);
eq2_9:= RLeq = %[1][1]
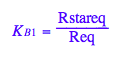
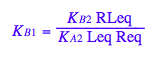
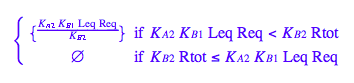
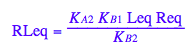
Check for mistakes in the equilbrium constants formulas:
Express Req using last equation left
eq2_3;
% | eq2_9;
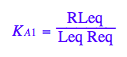
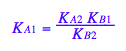
Nice!!! We arrived at the expected identity as one of the constants is dependent on the others!
Enter all equilibrium concentrations in equations for total concentrations:
eq2_1;eq2_2;
![]()
![]()
Substitute all receptor forms:
eq2_1;
% | eq2_7;
% | eq2_8;
% | eq2_9;
eq2_10:= %:
![]()
![]()
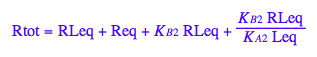
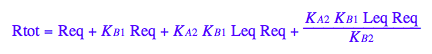
Express Req
solve(eq2_10,Req);
%[1];
eq2_11:= Req = %
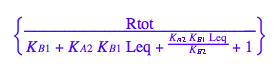
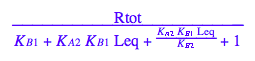

Substitute all ligand forms and Req
eq2_2;
% | eq2_7;
% | eq2_9;
% | eq2_11;
eq2_12:= %:
![]()
![]()

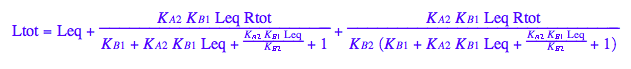
Solve for Leq
solutions2_12:=solve(eq2_12,Leq)

Extract solutions
eq2_13:= solutions2_12[i,1] $ i=1..nops(solutions2_12);
nops(%)
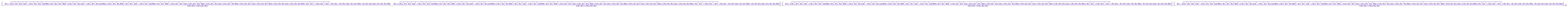
![]()
Is 1st solution a combination of 2nd and 3rd?
solution1:=eq2_13[1]; // a sequence of roots
solution2:=eq2_13[2][1]; // extract equation out of a sequence
solution3:=eq2_13[3][1]; // extract equation out of a sequence
if solution2 in solution1
then print(Unquoted,"First set of roots contains the second root.");
else print(Unquoted,"First set of roots DOES NOT contain the second root!");
end_if;
if solution3 in solution1
then print(Unquoted,"First set of roots contains the third root.");
else print(Unquoted,"First set of roots DOES NOT contain the third root!");
end_if;
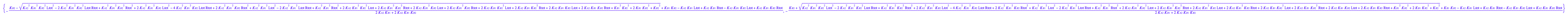
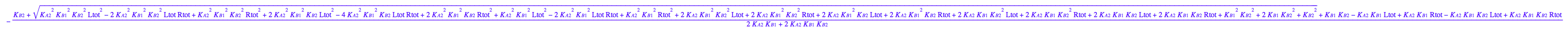
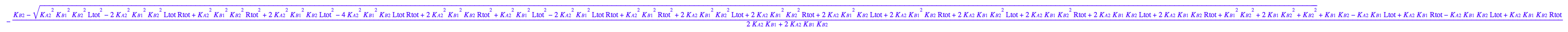
First set of roots contains the second root.
First set of roots contains the third root.
Check correctness of the solutions by substitution into original equation solved:
Check first root
test1:= eq2_12 | Leq=solution2;
normal(%);

![]()
Check second root
test2:= eq2_12 | Leq=solution3;
normal(%);

![]()
Both solutions are correct.
3. Define functions for equilibrium concentrations
Define functions for plotting and analysis
Choose one solution (tested for being meaningful later)
eq3_1:= Leq = solution3;
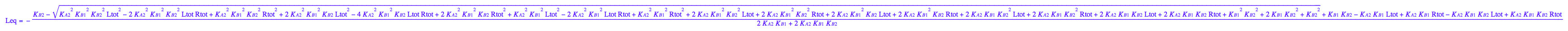
Express dependent constant:
eq2_3;
% | eq2_9;
solve(%,K_A_2);
%[1];
eq3_2:= K_A_2= %
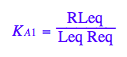
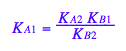
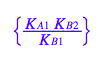
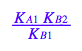
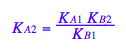
Remove dependent constant from all equations
eq3_1;
eq3_4:= eq3_1 | eq3_2;
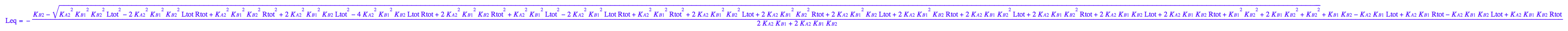
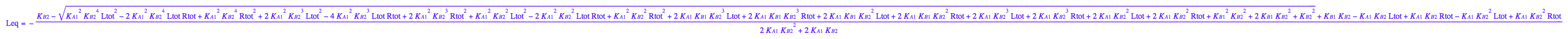
eq2_11;
eq3_5:= eq2_11 | eq3_4 | eq3_2;


eq2_9;
eq3_6:= % | eq3_4 | eq3_5 | eq3_2
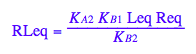

eq2_8;
eq3_7:= % | eq3_6 | eq3_4 | eq3_2;
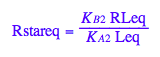

eq2_7;
eq3_8:= % | eq3_6;
![]()

Summary of equations for all species
Independent parameters:
Rtot, Ltot, K_A_1, K_B_1, K_B_2
![]()
Dependent constant
eq3_2
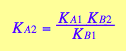
[L]
eq3_4
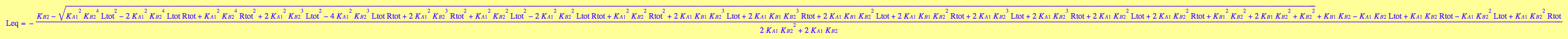
[R]
eq2_11

[RL]
eq2_9
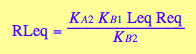
[R*]
eq2_8 | eq2_9
![]()
[R*L]
eq2_7 | eq2_9
![]()
Generate functions
fLeq:= (Rtot, Ltot, K_A_1, K_B_1, K_B_2) --> eq3_4[2]
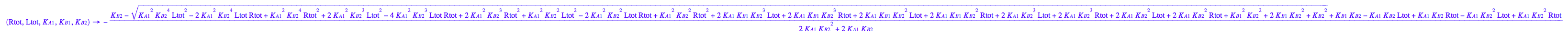
fReq:= (Rtot, Ltot, K_A_1, K_B_1, K_B_2) --> eq3_5[2]

fRLeq:= (Rtot, Ltot, K_A_1, K_B_1, K_B_2) --> eq3_6[2]

fRstareq:= (Rtot, Ltot, K_A_1, K_B_1, K_B_2) --> eq3_7[2]

fRstarLeq:= (Rtot, Ltot, K_A_1, K_B_1, K_B_2) --> eq3_8[2]

4. Test if solution is meaningful
Set some realistic values for constants:
Total_R:=1e-3;
Total_L:=0.5e-3;
Ka1:=1e7;
Kb1:=1/1;
Kb2:=0.1;
![]()
![]()
![]()
![]()
![]()
Test that all equilibrium concentrations are positive values:
if (fLeq(Total_R, Total_L, Ka1, Kb1, Kb2)>0 and
fReq(Total_R, Total_L, Ka1, Kb1, Kb2)>0 and
fRLeq(Total_R, Total_L, Ka1, Kb1, Kb2)>0 and
fRstareq(Total_R, Total_L, Ka1, Kb1, Kb2)>0 and
fRstarLeq(Total_R, Total_L, Ka1, Kb1, Kb2)>0
)
then
print(Unquoted,"Solution is meaningful.");
else
print(Unquoted,"WARNING!!!!");
print(Unquoted,"Solution is NOT meaningful: some concentrations become negative!");
end_if
Solution is meaningful.
5. Check whether the solution satisfies all initial equation and conditions
Here are all original independent
equations:
eq2_1;eq2_2;eq2_3;eq2_4;eq2_5;eq2_6;
![]()
![]()
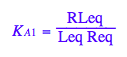
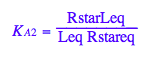
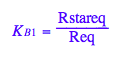
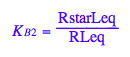
Test eq2_1
eq2_1;
% | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_8;
normal(%)
![]()
![]()
Test eq2_2
eq2_2;
% | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_8;
normal(%)
![]()

![]()
Test eq2_3
eq2_3;
% | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_8;
normal(%)
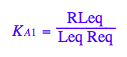
![]()
![]()
Test eq2_4
eq2_4;
% | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_8 | eq3_2;
normal(%)
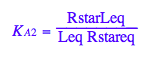
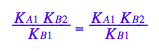
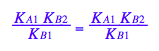
Test eq2_5
eq2_5;
% | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_8;
normal(%)
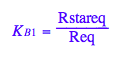
![]()
![]()
Test eq2_6
eq2_6;
% | eq3_4 | eq3_5 | eq3_6 | eq3_7 | eq3_8;
normal(%)
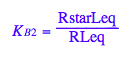
![]()
![]()
The found solution satisfies all original equations
5. Save results on disk for future use
(you can retrieve them later by executing: fread(filename,Quiet))
ProjectName
![]()
eq2_7;eq2_8;eq2_9;eq2_10;eq2_11;
![]()
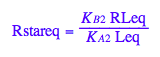
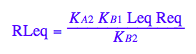
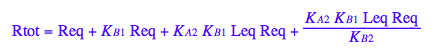

Generate Rtot=f([L]) equation for testing numerical routines against analytical solutions
eq2_2;
% | eq2_7;
% | eq2_9;
solve(%,Req);
%[1][1];
eq5_1:= Req = %;
![]()
![]()

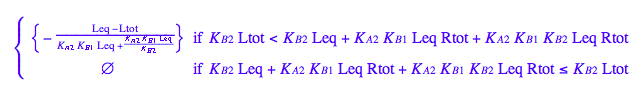
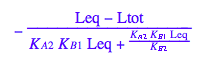
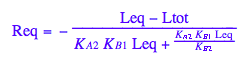
eq2_1;
% | eq2_7 | eq2_8 | eq2_9 | eq5_1 ;
% | eq3_2; // get rid of dependent KA2
eq5_2:= %:
![]()
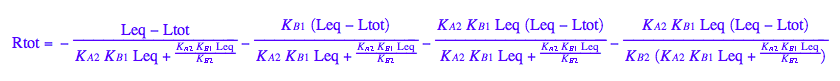
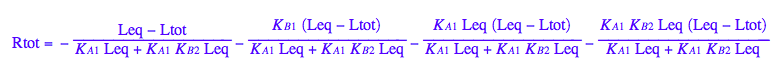
Here are solutions we want to save.
Reassign names
Rtot_U_R_RL:=eq5_2;
Leq_U_R_RL:=eq3_4:;
Req_U_R_RL:=eq3_5:;
RLeq_U_R_RL:=eq3_6:;
Rstareq_U_R_RL:=eq3_7:;
RstarLeq_U_R_RL:=eq3_8:;
KA2_U_R_RL:=eq3_2:;
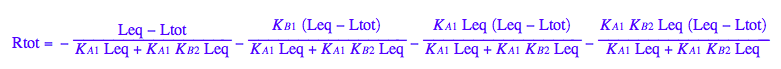
Reassign function names
fLeq_U_R_RL:=fLeq:
fReq_U_R_RL:=fReq:
fRLeq_U_R_RL:=fRLeq:
fRstareq_U_R_RL:=fRstareq:
fRstarLeq_U_R_RL:=fRstarLeq:
ProjectName
![]()
filename:=CurrentPath.ProjectName.".mb";
write(filename,Rtot_U_R_RL,Leq_U_R_RL, Req_U_R_RL, RLeq_U_R_RL, Rstareq_U_R_RL, RstarLeq_U_R_RL, KA2_U_R_RL,
fLeq_U_R_RL,fReq_U_R_RL,fRLeq_U_R_RL,fRstareq_U_R_RL,fRstarLeq_U_R_RL)
![]()
Conclusions
1. I derived analytical solutions for the U_R_RL system
2. I also derived and saved Rtot=f([L]) for numerical solution