Contents
Testing of the U_RL model
close all clear all
Here you will find all figures
figures_folder='U_RL_testing_figures';
TEST COMPUTATION OF EQUILIBRIUM POPULATIONS
Set some meaningful parameters
Rtotal=1e-3; % Receptor concentration, M LRratio_array=[0 : 0.02 : 1.45]; % Array of L/R K_a_A=1e6; % Binding affinity constant K_a_B=4; % Isomerization constant % Set appropriate options for the model (see model file for details) model_numeric_solver='analytical' ; model_numeric_options='none';
Important option here is TolX that sets termination tolerance on free ligand concentation in molar units. With our solution concentrations in 1e-3 range TolX should be set to some 1e-9.
Compute arrays for populations and plot
concentrations_array=[]; for counter=1:length(LRratio_array) % compute [concentrations species_names] = equilibrium_thermodynamic_equations.U_RL_model(... Rtotal, LRratio_array(counter), K_a_A, K_a_B,... model_numeric_solver, model_numeric_options); % collect concentrations_array = [concentrations_array ; concentrations]; end
Plot
Figure_title= 'U-RL model'; X_range=[0 max(LRratio_array)+0.1 ]; % extend X just a bit past last point Y_range=[ ]; % keep automatic scaling for Y % display figure figure_handle=equilibrium_thermodynamic_equations.plot_populations(... LRratio_array, concentrations_array, species_names, Figure_title, X_range, Y_range); % save it results_output.output_figure(figure_handle, figures_folder, 'Concentrations_plot');
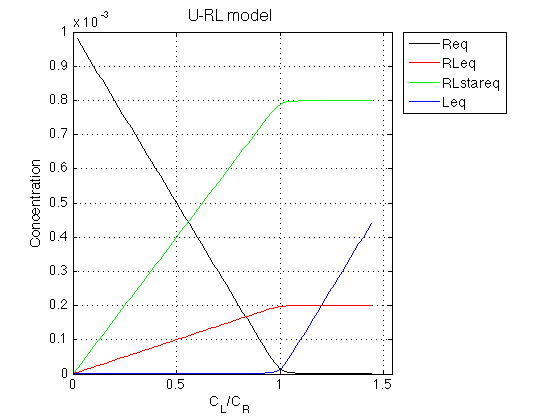
Observations The result is exactly what we expect from this model.
As ligand is added, the ligand-receptor complex is formed species is formed at a constant ratio between the two forms, RL and RL*
Conclusion
The model equilibrium_thermodynamic_equations.U_RL_model() works well.