Testing of the 1D line shape B-macro model (microscopic constants)
Evgenii Kovrigin, 12/27/2015
Contents
- Compute equilibruim populations using B-micro model
- Compute the same populations using microscopic constants in B-macro model
- Create the NMR line shape dataset
- 1. Free receptor
- 2. Saturated receptor
- 3. Equal R:RL:RL2 = 1:2:1
- 4. Dilution of the complex
- 5. Fast exchange in A1 for R:RL:RL2 = 1:2:1
- 6. Fast exchange in A2 for R:RL:RL2 = 1:2:1
Plan
- Set microscopic constants and calculate equilibrium populations using B-micro model with identical sites (parallel paths have the same constant)
- Convert microscopic constants into macroscopic ones the way B_macro_model_1D.m did and use B-macro model to calculate the same populations
- Calculate line shapes with different settings of the rate constants.
close all clear all
Here you will find all figures
figures_folder='B_macro_testing_figures';
Compute equilibruim populations using B-micro model
NOTE: Use microscopic constants and B-micro model assuming identical sites
Rtotal=1e-3; % Receptor concentration, M LRratio_array=[0 : 0.02 : 3]; % Array of L/R K_a_A1_micro=1e4; K_a_A2_micro=1e4; % Set parallel paths to identical constants K_A_1_I = K_a_A1_micro; K_A_1_I_I = K_a_A1_micro; K_A_2_I_I = K_a_A2_micro; % Set appropriate options for the model (see model file for details) model_numeric_solver='fminbnd' ; model_numeric_options=optimset('Diagnostics','off', ... 'Display','off',... 'TolX',1e-9,... 'MaxFunEvals', 1e9);
The important option here is "TolX" that sets termination tolerance on free ligand concentation in molar units. With our solution concentrations in 1e-3 range TolX should be set to some 1e-9.
Compute arrays for populations and plot
concentrations_array=[]; for counter=1:length(LRratio_array) % compute [concentrations species_names] = equilibrium_thermodynamic_equations.B_micro_model(... Rtotal, LRratio_array(counter),... K_A_1_I, K_A_1_I_I, K_A_2_I_I,... model_numeric_solver, model_numeric_options); % collect concentrations_array = [concentrations_array ; concentrations]; end
Plot
Figure_title= 'B-micro model'; X_range=[0 max(LRratio_array)+0.1 ]; % extend X just a bit past last point Y_range=[ ]; % keep automatic scaling for Y % generate Rtotal array Rtotal_array = ones(1, length(LRratio_array))*Rtotal; % display figure figure_handle=equilibrium_thermodynamic_equations.plot_populations(... Rtotal_array, LRratio_array, concentrations_array, species_names, Figure_title, X_range, Y_range); % save it results_output.output_figure(figure_handle, figures_folder, 'Concentrations_plot');
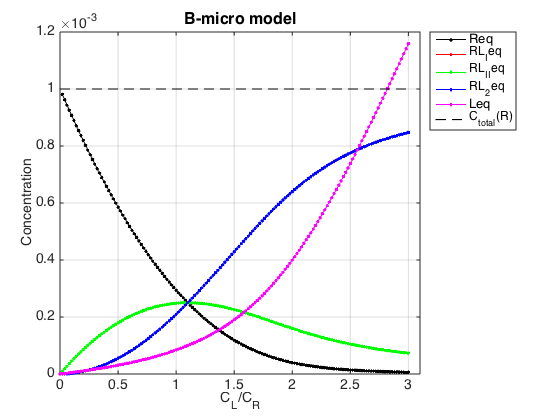
Compute the same populations using microscopic constants in B-macro model
Calculate macroscopic constants using the same equations as in B_macro_model_1D.m. Goal: test whether the conversion to macroscopic constants was done correctly
K_a_A1 = 2*K_a_A1_micro K_a_A2 = K_a_A2_micro/2 concentrations_array=[]; for counter=1:length(LRratio_array) % compute [concentrations species_names] = equilibrium_thermodynamic_equations.B_macro_model(... Rtotal, LRratio_array(counter), K_a_A1, K_a_A2,... model_numeric_solver, model_numeric_options); % collect concentrations_array = [concentrations_array ; concentrations]; end
K_a_A1 =
20000
K_a_A2 =
5000
Plot
Figure_title= 'B-macro model'; X_range=[0 max(LRratio_array)+0.1 ]; % extend X just a bit past last point Y_range=[ ]; % keep automatic scaling for Y % generate Rtotal array Rtotal_array = ones(1, length(LRratio_array))*Rtotal; % display figure figure_handle=equilibrium_thermodynamic_equations.plot_populations(... Rtotal_array, LRratio_array, concentrations_array, species_names, Figure_title, X_range, Y_range); % save it results_output.output_figure(figure_handle, figures_folder, 'Concentrations_plot');
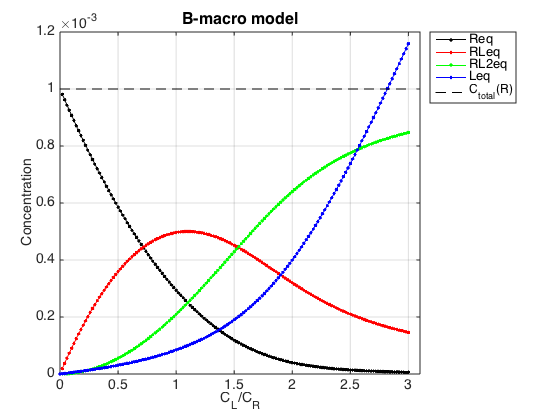
Yes, the conversion to macroscopic constants was correct.
Create the NMR line shape dataset
Create data object for 1D NMR line shapes
test1=NMRLineShapes1D('Simulation','B'); test1.set_active_model('B-macro-model', model_numeric_solver, model_numeric_options) test1.show_active_model() % to look up necessary parameters of the model
ans = Active model: Model 17: "B-macro-model" Model description "1D NMR line shape for the B-macro model with two identical sites using MICROSCOPIC constants, no temperature/field dependence, fminbnd solver" Model handle: line_shape_equations_1D.B_macro_model_1D Current solver: fminbnd Model parameters: 1: Rtotal 2: LRratio 3: log10(K_A1) 4: log10(K_A2) 5: k_2_A1 6: k_2_A2 7: w0_R 8: w0_RL 9: w0_RL2 10: FWHH_R 11: FWHH_RL 12: FWHH_RL2 13: ScaleFactor 14: LRcorrection
Set range for X to extend beyond resonances
w_min= -300; w_max= 500; datapoints=100; % Does not matter much because smooth curve is anyway calculated test1.set_X(linspace(w_min, w_max, datapoints)); test1.X_range=[w_min w_max]; % this sets display range in the plots test1.Y_range=[0 18e-5]; % this sets display range in the plots
Calculate ideal data: set noise RMSD to 0 for both X and Y
X_RMSD=0; Y_RMSD=0; % *Prepare TotalFit session for easy display of many graphs* storage_session=TotalFit('All_tests');
1. Free receptor
Use the same thermodynamic parameters as above. Add spectral and kinetic parameters:
LRratio = 0.001 ; Log10_K_A1 = log10(K_a_A1_micro); Log10_K_A2 = log10(K_a_A2_micro); k_2_A1 = 1; % dissociation rate constant of the complex, 1/s k_2_A2 = 1; % dissociation rate constant of the receptor dimer, 1/s w0_R = -200; % NMR frequency of the free receptor R, 1/s w0_RL = 200; % NMR frequency of the dimer R2, 1/s w0_RL2 = 400; % NMR frequency of the bound complex RL, 1/s FWHH_R = 10; % line width at half height of the peak of R, 1/s FWHH_RL = 10; % line width at half height of the peak of R2, 1/s FWHH_RL2 = 10; % line width at half height of the peak of RL, 1/s ScaleFactor = 1; % a multiplier for spectral amplitude (used only when fitting data) LRcorrection = 1; % default: no correction % plot line shapes parameters=[ Rtotal LRratio Log10_K_A1 Log10_K_A2 k_2_A1 k_2_A2 ... w0_R w0_RL w0_RL2 FWHH_R FWHH_RL FWHH_RL2 ScaleFactor LRcorrection]; test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure_handle=test1.plot_simulation('Free R'); results_output.output_figure(figure_handle, figures_folder, 'Slow_free_R'); % set aside for plotting later storage_session.copy_dataset_into_array(test1);
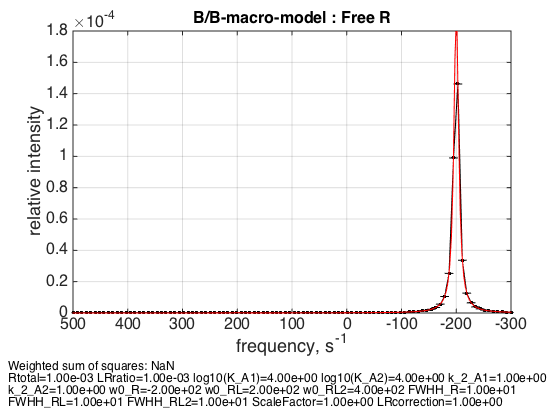
As expected, free receptor signal is centered at w0_R frequency
2. Saturated receptor
% Use the same thermodynamic parameters as above. Add spectral and kinetic parameters: LRratio = 10 ; % plot line shapes parameters=[ Rtotal LRratio Log10_K_A1 Log10_K_A2 k_2_A1 k_2_A2 ... w0_R w0_RL w0_RL2 FWHH_R FWHH_RL FWHH_RL2 ScaleFactor LRcorrection]; test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure_handle=test1.plot_simulation('Saturated R'); results_output.output_figure(figure_handle, figures_folder, 'Slow_saturated_R'); % set aside for plotting later storage_session.copy_dataset_into_array(test1);
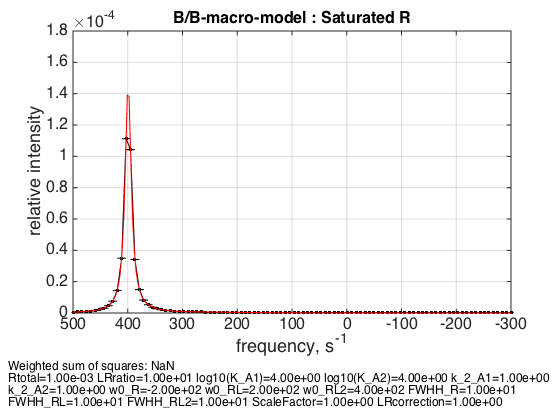
Peak appropriately appears at the w0_RL2 frequency
3. Equal R:RL:RL2 = 1:2:1
LRratio = 1.1; % plot line shapes parameters=[ Rtotal LRratio Log10_K_A1 Log10_K_A2 k_2_A1 k_2_A2 ... w0_R w0_RL w0_RL2 FWHH_R FWHH_RL FWHH_RL2 ScaleFactor LRcorrection]; test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure_handle=test1.plot_simulation('R:RL:RL2 = 1:2:1'); results_output.output_figure(figure_handle, figures_folder, 'middle_point'); % set aside for plotting later storage_session.copy_dataset_into_array(test1);
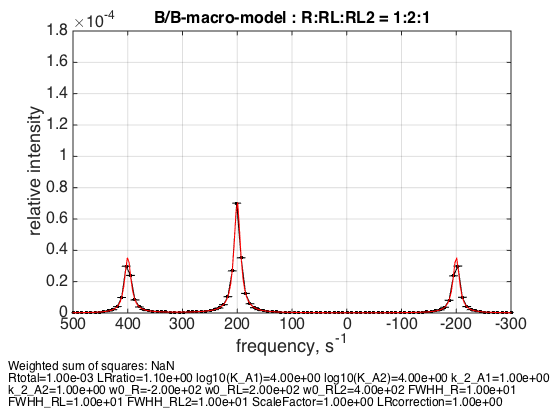
I picked this point because there is exact 1:2:1 ratio between populations at this specific receptor concentration.
4. Dilution of the complex
Rtotal=1e-4; % plot line shapes parameters=[ Rtotal LRratio Log10_K_A1 Log10_K_A2 k_2_A1 k_2_A2 ... w0_R w0_RL w0_RL2 FWHH_R FWHH_RL FWHH_RL2 ScaleFactor LRcorrection]; test1.Y_range=[0 1e-5]; % this sets display range in the plots test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure_handle=test1.plot_simulation('middle point diluted'); results_output.output_figure(figure_handle, figures_folder, 'middle_point_diluted'); % set aside for plotting later storage_session.copy_dataset_into_array(test1);
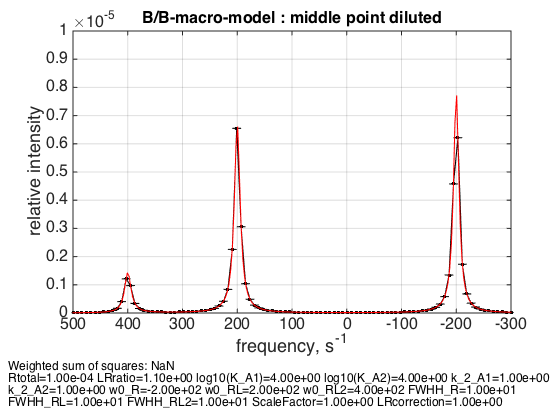
Dilution appropriately shifts equilibrium back.
5. Fast exchange in A1 for R:RL:RL2 = 1:2:1
Rtotal=1e-3; LRratio = 1.1; k_2_A1=1000; k_2_A2=1; % plot line shapes parameters=[ Rtotal LRratio Log10_K_A1 Log10_K_A2 k_2_A1 k_2_A2 ... w0_R w0_RL w0_RL2 FWHH_R FWHH_RL FWHH_RL2 ScaleFactor LRcorrection]; test1.Y_range=[0 10e-5]; % this sets display range in the plots test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure_handle=test1.plot_simulation('fast A1 at middle point'); results_output.output_figure(figure_handle, figures_folder, 'middle_point_fast_A1'); % set aside for plotting later storage_session.copy_dataset_into_array(test1);
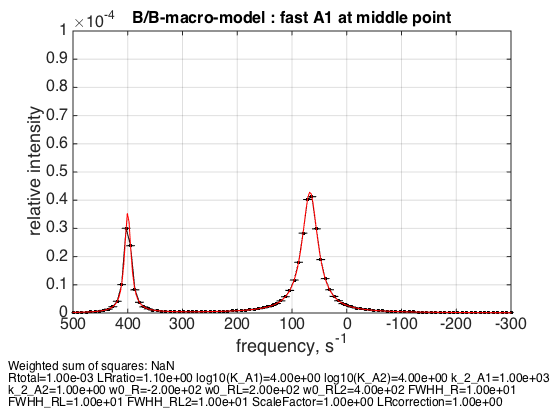
The weighted average peak appears at 1/3 : 2/3 distance between RL and R as expected.
6. Fast exchange in A2 for R:RL:RL2 = 1:2:1
Rtotal=1e-3; LRratio = 1.1; k_2_A1=1; k_2_A2=1000; % plot line shapes parameters=[ Rtotal LRratio Log10_K_A1 Log10_K_A2 k_2_A1 k_2_A2 ... w0_R w0_RL w0_RL2 FWHH_R FWHH_RL FWHH_RL2 ScaleFactor LRcorrection]; test1.Y_range=[0 10e-5]; % this sets display range in the plots test1.simulate_noisy_data(parameters, X_RMSD, Y_RMSD); figure_handle=test1.plot_simulation('fast A2 at middle point'); results_output.output_figure(figure_handle, figures_folder, 'middle_point_fast_A2'); % set aside for plotting later storage_session.copy_dataset_into_array(test1);
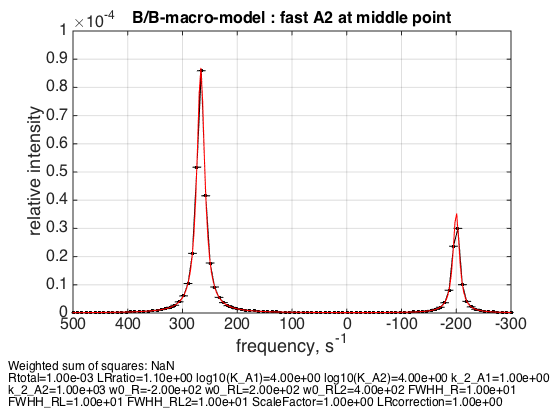
The weighted average peak between RL2 and RL appears at 2/3 : 1/3 distance as expected.
Compare these results to the LineShapeKin Simulation reports with B-micro
IDAP/Mathematical_models/NMR_line_shape_models/1D/B/LineShapeKin_Simulation_reference/Example_simulation
- Tests 1, 2, and 3 : Simulate setup123 slowA1_slowA2
- Tests 3 and 4: Simulate setup34 slowA1_slowA2
- Test 5 : Simulate setup5 fastA1_slowA2
- Test 6 : Simulate setup6 slowA1_fastA2
All tests appeared identical to LineShapeKin Simulation calculations