
In this model the free ligand dimerizes. Only a monomeric ligand is capable to bind the receptor. This mechanism is also relevant to cases of aggregation, where weak, fast-off dimerization promotes slow irreversible aggregation of the ligand. The model is complementary to U_R model: they form forward/reverse titration pair.
This is a very simple model, essentially identical to U-model, because kinetics of ligand dimerization has no effect on line shapes. The only effect is through thermodynamic mass action law that dictates different monomer concentrations during titrations.
Simulate setup U_L2_As_Bs
report: summary_report
Fit with 2-site model:

Optimum norm: 6.86e-04
[1] Kd: 1.62209e-08 +/- 3.0e-09
[2] Koff: 9.03659e-01 +/- 1.6e-02
[3] Scale Factor: 2.02189e+00 +/- 1.5e-03
Fitted Ka=1/Kd=0.61e+07 with 1e8 preset value
Koff= 0.9/s vs 1/s pre-set.
Conclusion:
Apparent affinity is reduced approx. 2-fold by competing dimerization process.
Simulate setup U_L2_Af_Bs
report: summary_report
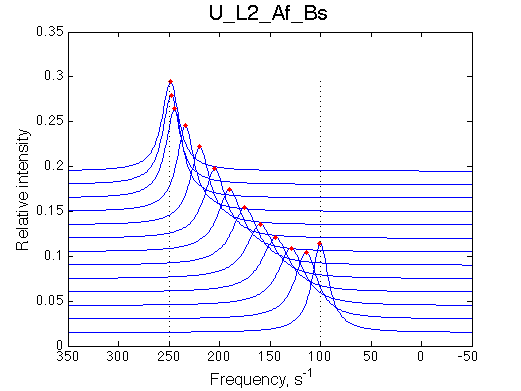

Optimum norm: 6.03e-03
[1] Kd: 5.22524e-07 +/- 2.4e-08
[2] Koff: 5.27700e+02 +/- 8.0e+00
[3] Scale Factor: 2.03559e+00 +/- 4.6e-03
Fitted Ka=1/Kd=2e+06 with 1e6 preset value
Koff= 530/s vs 500/s pre-set.
Conclusion:
It is counterintuitive but fitting with the 2-site model produces 2x tighter apparent affinity.
1. Fitting of both slow and fast binding examples with the 2-site model produces very good fit.
2. In case of tight, slow-off binding, the apparent affinity is reduced approx. 2-fold.
3. In case of weak, fast-off binding, the apparent affinity is increased approx. 2-fold.
Back to LineShapeKin Simulation Tutorial