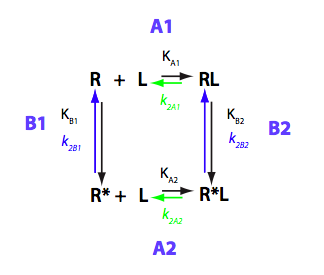

This is the model for the ligand-induced population shift. We may think of, say, R* being a major conformer yet less capable of binding ligand. Then RL complex will be more populated in the bound state than R*L. Thus adding a ligand we induce a shift in R-R* isomerization equilibrium.
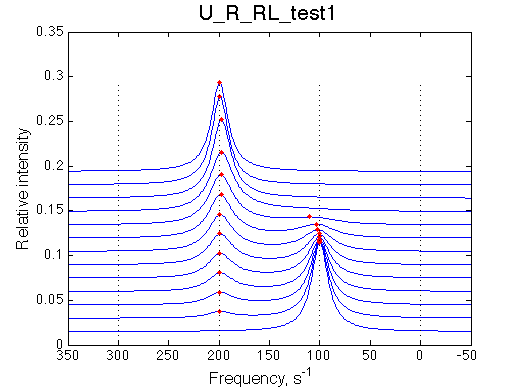
By choosing appropriate constants we 'turn off' parts of the model. We choose slow rates to see individual peaks well.
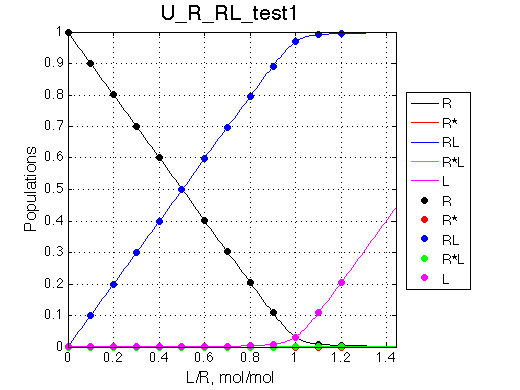
No R* and R*L |
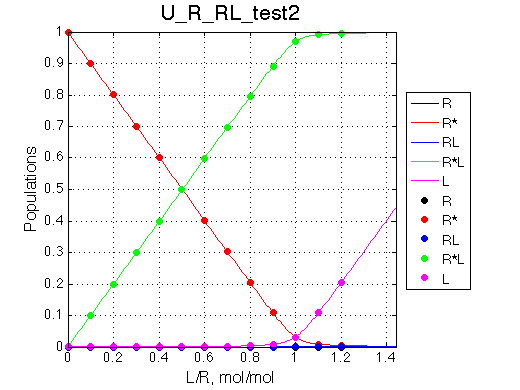
No R and RL |
| Simulate setup_implementation_test U_R_RL_test1 | Simulate setup_implementation_test U_R_RL_test2 |
 |
 |
 |
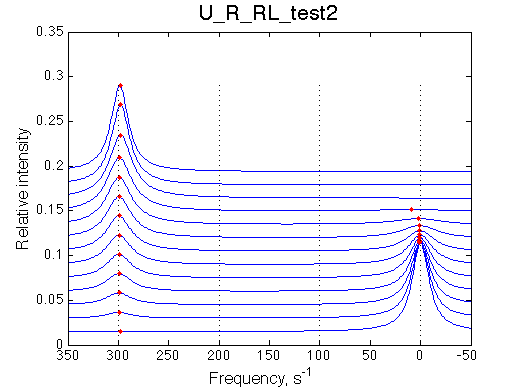 |
We are able to remove parts of the model and it behaves as expected.
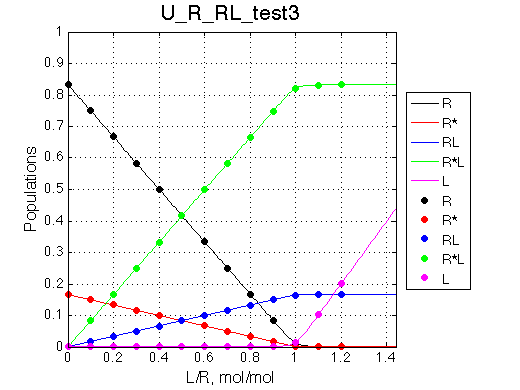
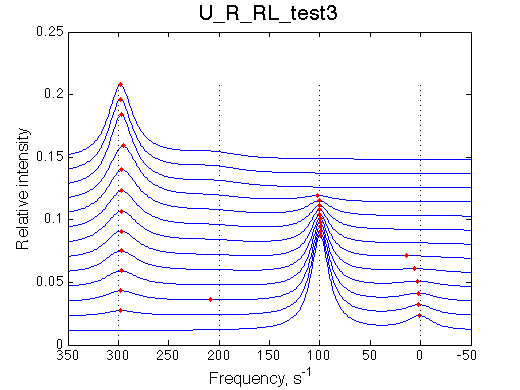
We want to have R as a major conformer in free receptor while R*L will be major species of the complex.
Simulate setup_implementation_test U_R_RL_test3
 |
 |
It works.
It works.
Back to LineShapeKin Simulation Tutorial