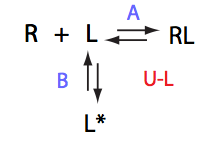
In this model an intramolecular isomerization occurs in the ligand molecule. Only one of the two isomers is capable to appreciably bind the receptor. This is a mechanism with a preexisting conformational equilibrium.
This model may also be thought of as a reverse titration of the U_R system so that the R and L in U_R exchange places: now the receptor is unlabeled and used as L while former ligand is now labeled and observed so it treated as R (NMR-observable species).

We will explore a couple cases in this system. First, I set up simulation for the slow isomerization process with 3:1 ratio in favor of L* and micromolar binding of L with the fast off-rate.
NOTE: Kinetics of L-L* rearrangement cannot affect the lineshapes because kinetic constants for L isomerization do not enter Bloch-McConnell equations. Only if kinetics is so slow that equilibrium is NOT established during NMR sample preparation, then it will impact on line shapes but that case must be treated as non-equilibrium system! If we have off-rate for the B process to be 10 s-1 then in less than a second we will have the equilibrium established.
Simulate setup U_L_Af_Bs
Report: summary_report.
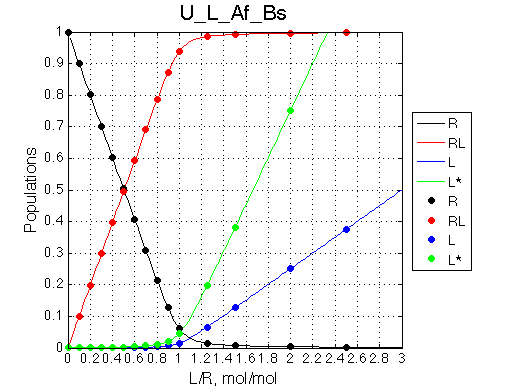
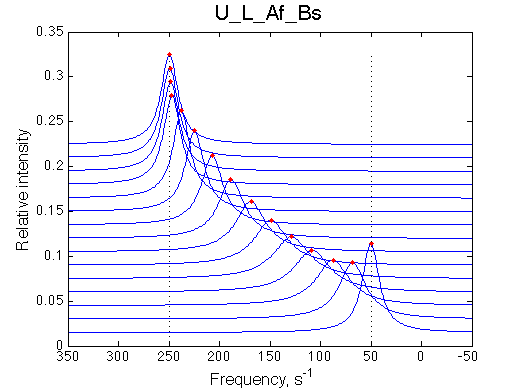
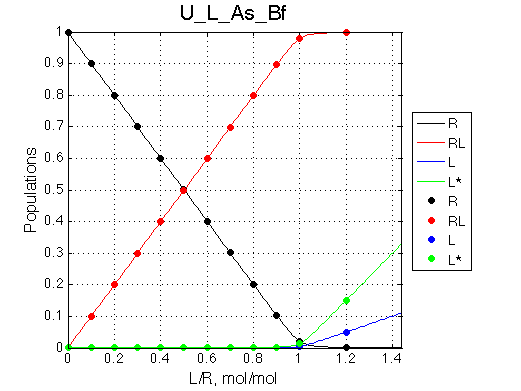
One of the most significant features of this model is that population of the free ligand is always split between L (blue symbols) and L* (green symbols) with the same ratio given by the equilibrium constant KB .
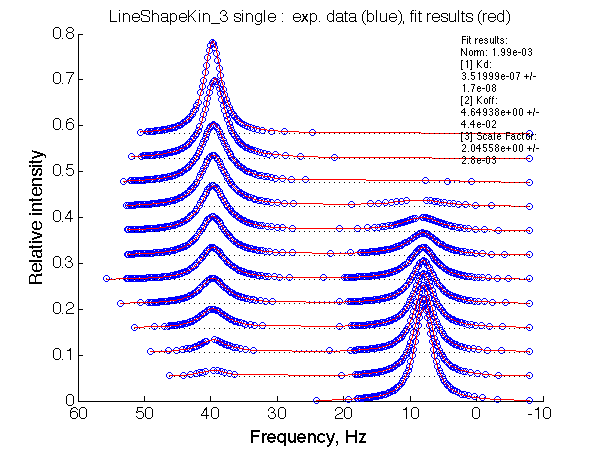
Let's fit this data with the two-site model (instructions):
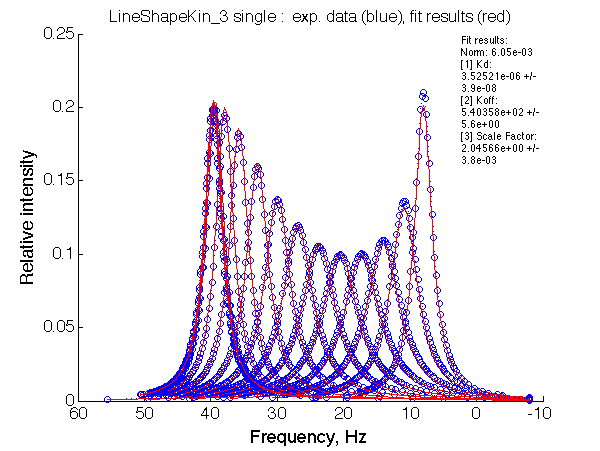
Fit is excellent. The apparent association constant Ka =1/Kd =0.29*106 . Thus isomerization in favor of non-binding form of L reduced affinity by (KB+1) according to the equation Kapp =KA/(1+KB). The spectra, however, don't give us any hint of presence of isomerization process in the ligand.
U_L with fast binding and slow isomerization of ligand is a situation reverse to the example 2 of U_R model (isomerization of receptor, which produces very clear indication of the 3-state exchange. Thus, when fast exchange regime is observed, it is important to perform binding studies both ways: adding an unlabeled ligand to labeled receptor and, opposite, unlabeled receptor to the labeled ligand.
Settings:
Simulate setup2 U_L_As_Bs
Simulate setup2 U_L_As_Bf
Report: summary_report
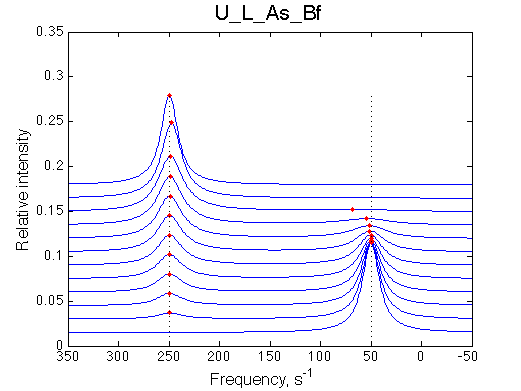
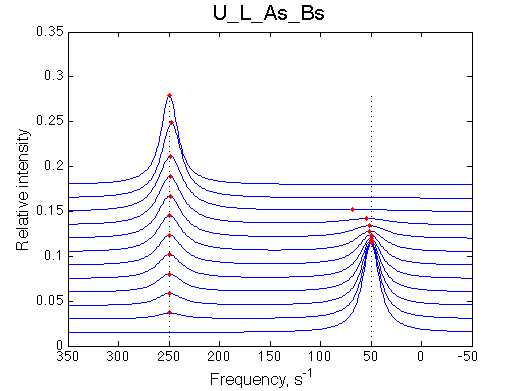
Fitting with the 2-site model gives, correspondingly:
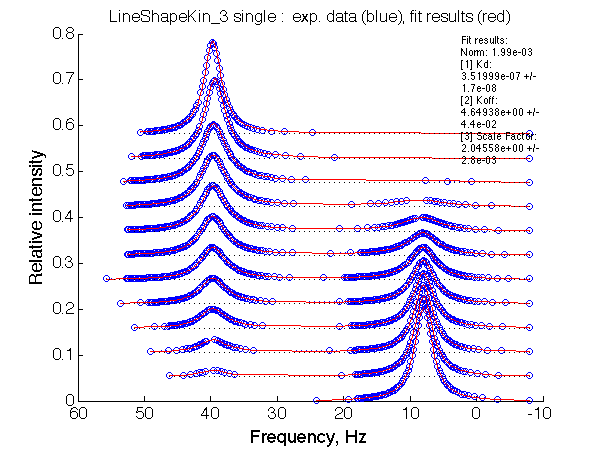
Fit is excellent, this data is not going to support any model better than 2-site exchange!
Affinity of interaction 1/3.5*107 = 2.9*106 M-1 obtained from a 2-state fit reflects the binding affinity scaled by the isomerization constant: Kapp =KA/(1+KB) = 107 /(1+3)=2.5*106 M-1 .
This situation is reverse to the Example 4 in the U_R model where titration of the labeled receptor with an unlabeled ligand does not allow to discern 2- and 3-site binding mechanisms if binding is slow.
However, if isomerization is slow then reversing roles of ligand and a receptor will yield slow exchange spectra where 3-site exchange may potentially be detected.
Overall, titration with the isomerizing species has no effect on line shapes due to the other (non-isomerizing) species.
U_L with fast binding and slow isomerization of ligand is a situation reverse to the example 2 of U_R model (isomerization of receptor, which produces very clear indication of the 3-state exchange. Thus, when fast exchange regime is observed, it is important to perform binding studies both ways: adding an unlabeled ligand to labeled receptor and, opposite, unlabeled receptor to the labeled ligand.
When ligand binding event has slow off-rate, the only manifestation of isomerization of NMR-unobservable component is that binding affinity is scaled down by the equilibrium constant of formation of the non-binding species. In addition, if isomerization is in fast exchange then the reverse titration (U_R, Example 3) cannot resolve the two processes. However, if isomerization is slow then observing isomerizing species while titrating in the other one will produce a signature pattern of the 3-site exchange (U_R, Example 4).
Back to LineShapeKin Simulation Tutorial