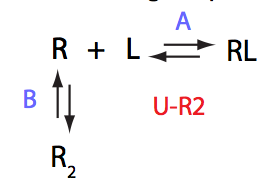

In this model the receptor molecule dimerizes. Only the monomer is capable to appreciably bind the ligand. This mechanism is also relevant to cases of aggregation, where weak, fast-off dimerization promotes slow irreversible aggregation of the receptor.
This is the simplest example. All species are in slow exchange, we will see three peaks. I set up simulation for the slow isomerization process with 3:1 ratio in favor of R* and nanomolar binding with very slow off-rate.
NOTE: ITC and smooth population curves will NOT work for changing concentration of the receptor!!!
Example_concentration_dependence/
Report: summary_report
slow dissociation |
fast dissociation |
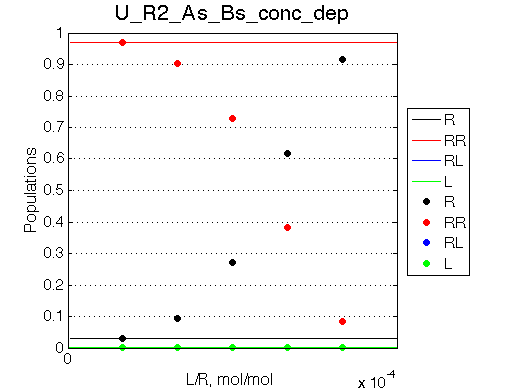
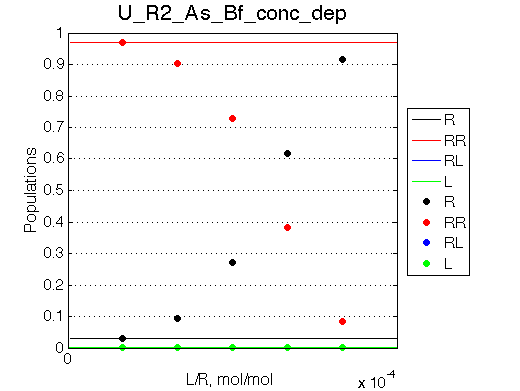
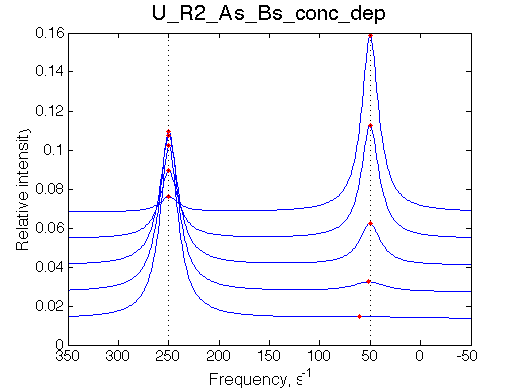
| Simulate setup_conc_dep U_R2_As_Bs_conc_dep | Simulate setup_conc_dep U_R2_As_Bf_conc_dep |
 |
 |
 |
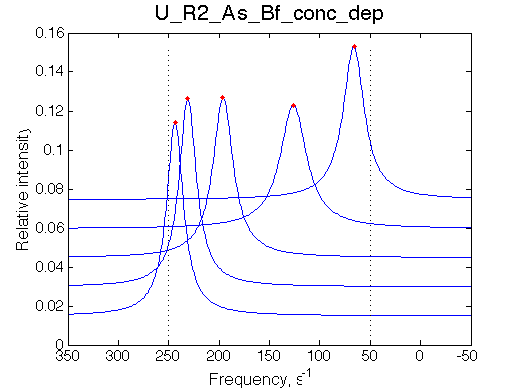 |
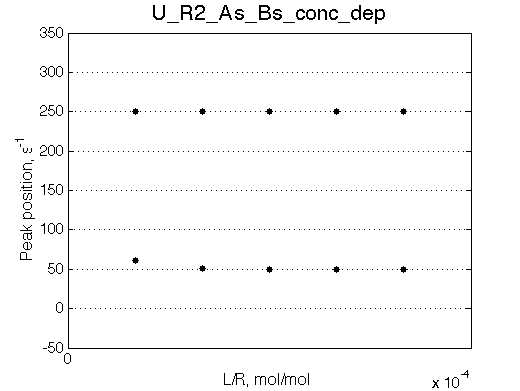 |
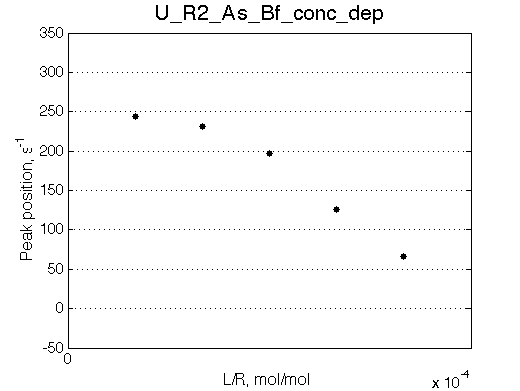 |
The model was reasonably reduced to the simple self-association model.
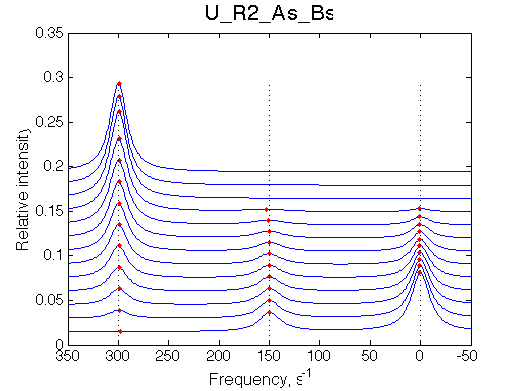
Simulate setup U_R2_As_Bs
report: summary_report
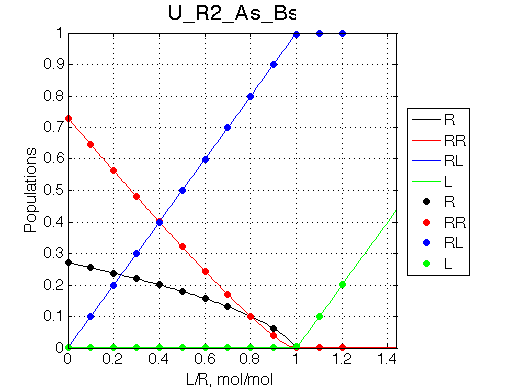
Simulate setup U_R2_Af_Bs
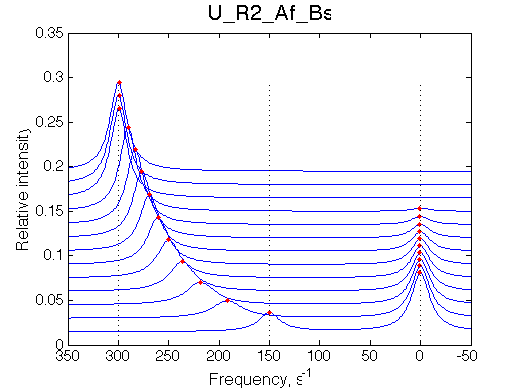
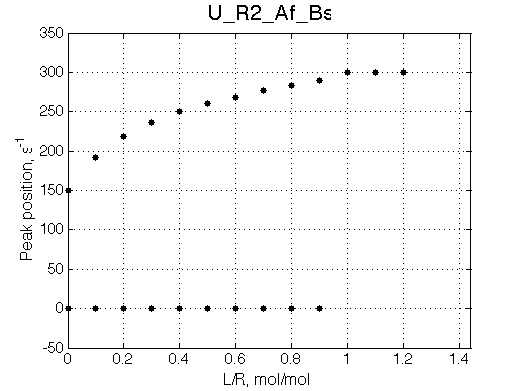
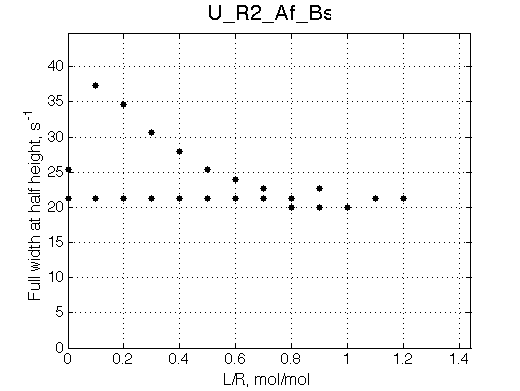
NOTE: This situation is VERY SIMILAR to behavior of U_R system. Use ITC profile to tell them apart.
Simulate setup U_R2_Af_Bf
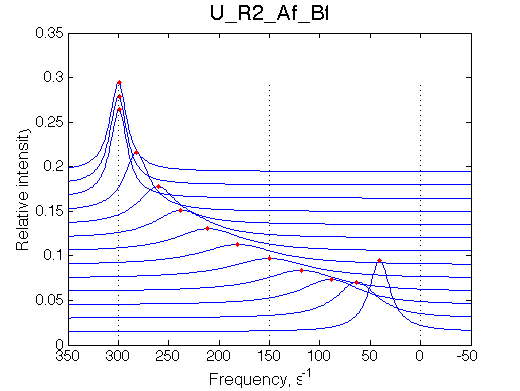
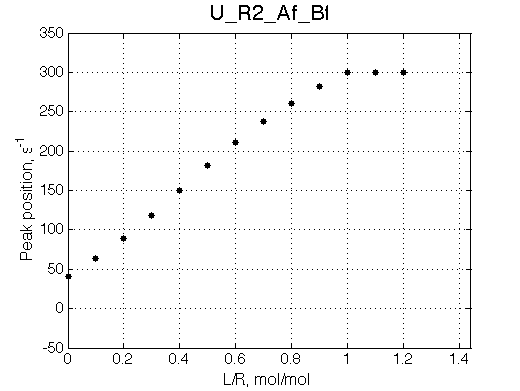
Note non-linearity of the FWHH curve shifted to 0.4 relatively to U_R model
The noted non-linearity makes the fit with the 2-site model fail miserably:
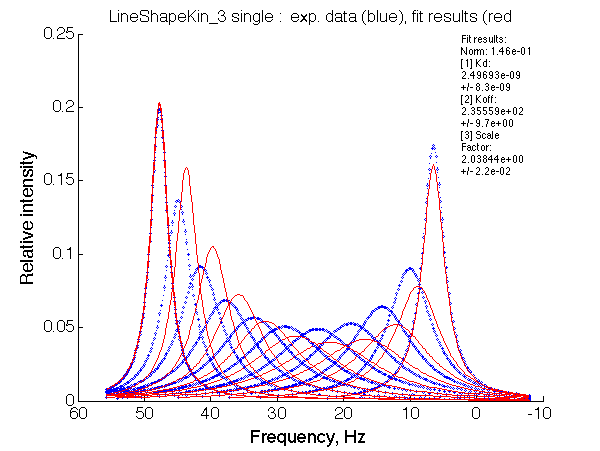
Such discrepancy between data and the fit is very remarkable, dissimilar to the U_R and, probably, to U_RL.
Simulate setup U_R2_As_Bf
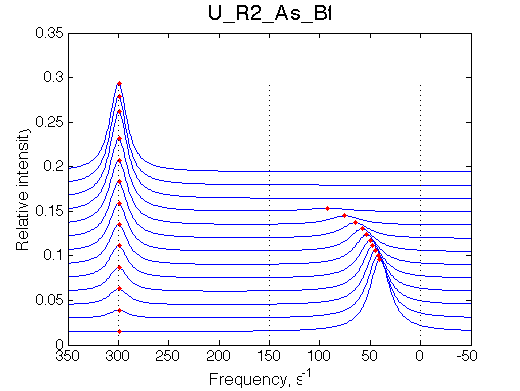
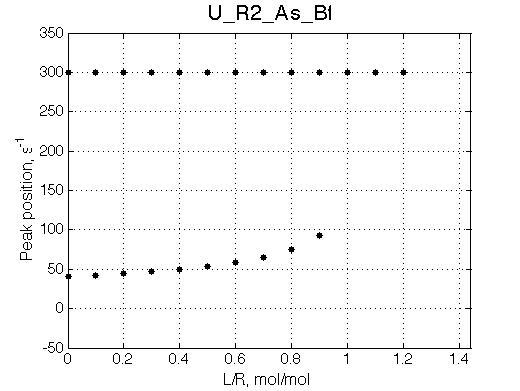
NOTE: This spectral appearance is VERY SIMILAR to U_RL model and U_R2L2
Back to LineShapeKin Simulation Tutorial